Our group investigates X-chromosome reactivation during the specification of the germline. This female-specific reprogramming pathway is a paradigm of epigenetic resetting and fine gene-regulation control. A better understanding of the mechanisms involved in gene expression regulation and epigenetic memory is crucial, especially in the very unique context of the germ line formation.
Epigenetic memory plays a determinant role in locking cell fates through inheritance of chromatin modifications such as DNA methylation and histone marks. This is paramount for the stability of gene expression programs, cell identity and functions. In mammals, the most drastic transition in cell fate coincides with the emergence of the germline in the embryo. It involves a profound epigenetic remodelling of the cells. Occurring in both sexes, germline reprogramming is essential for the specialization of reproductive cells, and therefore, for the perpetuation of the species. However, in females specifically, reactivation of the entire inactive X chromosome, an epigenetic hallmark of female somatic cells, accompanies the acquisition of germline identity. Incidentally, this leads to a biallelic expression of X-linked gene in the germline precursors of XX females compared to XY males, at the onset of gonadal sex determination. The biological impact and significance of this sex-specific epigenetic characteristics is unknown. However, the presence of a non-reactivated X chromosome could be detrimental to homologous chromosome pairing and segregation in meiosis, and could lead to maternally inherited sex-chromosome aneuploidies such as XO and XXY syndromes in humans.
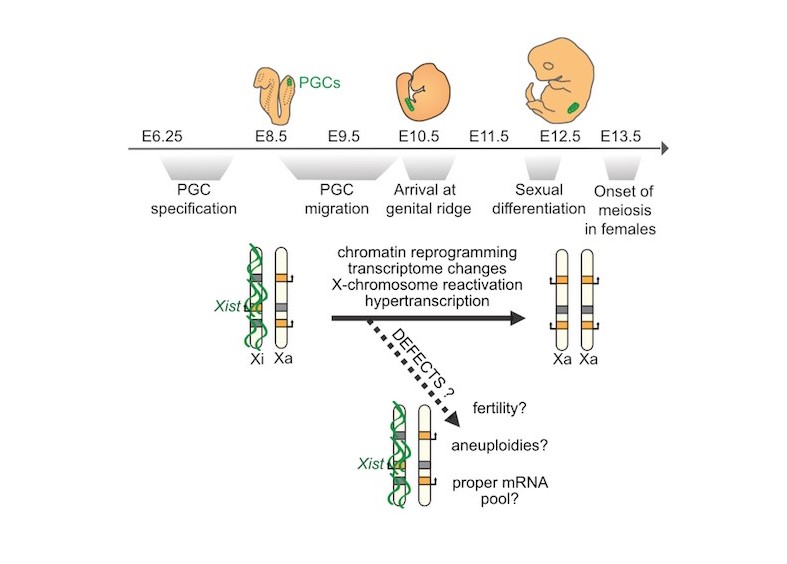
Our lab explores how sex differences in gene expression—particularly involving normal, aneuploid, and defective X-chromosome inactivation and reactivation—shape female reproduction and early development.

The path to reactivation, aquarelle, C. Roidor 2024
Who we are:


Who are you


Selection of publications :
-Temporal and regional X-linked gene reactivation in the mouse germline reveals site-specific retention of epigenetic silencing. Roidor C, Syx L, Beyne E, Raynaud P, Zielinski D, Teissandier A, Lee C, Walter M, Servant N, Chebli K, Bourc'his D, Surani MA, Borensztein M.Nat Struct Mol Biol. 2025 Jan 21. doi: 10.1038/s41594-024-01469-2. PMID: 39838109
-[Epigenetic reprogramming, germline and genomic imprinting]. Roidor C, Chebli K, Borensztein M.Med Sci (Paris). 2024 Dec;40(12):892-903. doi: 10.1051/medsci/2024177. Epub 2024 Dec 20.PMID: 39705560
-X-chromosome inactivation: a historic topic that's still hot. Moyano Rodriguez Y, Borensztein M.2023 Nov 15;150(22):dev202072. doi: 10.1242/dev.202072. Epub 2023 Nov 23.PMID: 37997921
-Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass. Borensztein M, Okamoto I, Syx L, Guilbaud G, Picard C, Ancelin K, Galupa R, Diabangouaya P, Servant N, Barillot E, Surani A, Saitou M, Chen CJ, Anastassiadis K, Heard E.Nat Commun. 2017 Nov 3;8(1):1297. doi: 10.1038/s41467-017-01415-5.PMID: 29101321
-Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N, Barillot E, Surani A, Chen CJ, Heard E.Nat Struct Mol Biol. 2017 Mar;24(3):226-233. doi: 10.1038/nsmb.3365. Epub 2017 Jan 30.PMID: 28134930
List of all publications



