A novel General Genome Interpretation Bioinformatics paradigm
for Precision Medicine
Motivation
From a scientific perspective, understanding our genome means being able to model the relationship between genotype and phenotype. This is one of the most ambitious currently open scientific challenges, and this achievement would be indeed groundbreaking for biology, genetics, medicine and agro-tech. For example, it could help identify in advance late onset genetic disorders, and lead to the design of treatments tailored to each patient's genome, complementing environmental and medical-history data to improve patient prognosis. These kinds of personalized approaches to medicine are still out of our scientific reach, and are called Precision Medicine1. In agro-tech, it could lead to the faster development of better crops, able to face the challenges posed by global warming and food access inequalities. Applied to cancer, it could bring a novel understanding of cancerogenesis, giving us the opportunity to devise highly specific cocktails of drugs or even engineer specific molecules to target each unique tumor.
Main objective
Genome Interpretation (GI) is the umbrella term for the genetic, statistic and computational attempts to further our understanding of how the mutations (variants) in our genome lead to the observed phenotypes 2,3, which can be quantitative (e.g., height, weight), qualitative (e.g., hair color, eye color) or diseases (e.g., sickle-cell anemia, Hungtinton’s disease) 4, 5 .My approach tackles GI from a novel perspective with respect to what has been done in genetics and bioinformatics so far, where classical, and sometimes simplistic statistical approaches have been used to model complex genetics mechanisms. To mark this difference with conventional GI attempts, I call this novel paradigm General Genome Interpretation (GenGI). GenGI consists of a data-driven framework of complex, yet biologically meaningful, problem-specific Deep Learning (DL) models, able to combine Neural Networks architectures with prior knowledge of biological processes directly in their architecture.
Core design philosophy
End-to-end DL models recently gained popularity thanks to the astonishing performance of 6. It is a design philosophy where a single, global, model learns all the steps between the raw input data and the desired final output predictions. In the GenGI context, this means that my DL framework will be able to take as input raw genomics data, such as Whole Genome or Exome Sequencing data (WGS, WES)7or other omics, and will directly provide phenotypic predictions as outputs, thus modeling the relationship between genotype and phenotype in a unique computation stream (end-to-end) 8. In our models we will thus consider each genomic sample as a whole, by:
1. Adopting a data-driven approach to let inheritance and disease etiology emerge from the data
2. Allowing nonlinear modeling of the observed variants and their interaction, considering phenotypes as complex emergent properties of the genotype
3. Developing ad hoc, tailor-made DL architectures for each unique GenGI task we tackle, integrating biological and task specific knowledge in them whenever it is available.
4. by extracting biological knowledge from the interpretation of the predictions with XAI techniques, ideally producing biological hypotheses about the inheritance mechanisms that could be experimentally tested.
I refer to GenGi as a paradigm, because I indeed envision it as a general framework of DL architectures able to tackle a wide spectrum of GI problems. In my research, I am indeed following the core design principles described above to address each specific problem in genetics, biology, agro-tech and cancer medicine we face, developing ad hoc solutions for every case.
Scientific Objectives (SO)
The GenGI tenets I described above can be concretely instantiated into a long term research project with multiple branches, each leading to a different SO. Each SO should ideally be assigned to a Ph.D. student or post-doc under my direct supervision. Each SO is independent from the others in terms of its final goal and its desired biological results, but they all share the underlying GenGI conceptual and methodological approach in terms of the DL/ML technologies used, allowing interaction and collaboration between the researchers involved, as well as technology transfer between them.
SO1: Disease risk prediction of human diseases (such as Inflammatory Bowel Disease (IBD))
SO2: Biologically Meaningful DL models for the wide-spectrum prediction of human phenotypes
SO3: DL Data fusion methods for multi-omics Pharmacogenomic discovery of anticancer drugs
SO4: Biologically Meaningful Sparsified DL models for multi-phenotypic prediction in A. thaliana
Even though each SO has its peculiarities, they can just be considered as different flavors of GenGI. In my vision, GenGI can indeed be framed within a spectrum of complexity, as shown in the Figure on the left. The narrow sense GenGI addresses 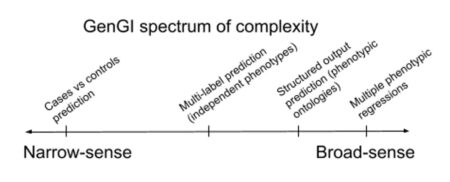 cases versus controls binary discrimination, while in the broad sense the goal is to predict multiple phenotypes for each sample. The papers on end-to-end GI that I have recently published 8,9,10,11 are the embryos from which I will expand this project. In particular, our Crohn’s disease (CD) case-control predictors8,9,10,11 belonged to the narrow sense GenGI, while Galiana, the multi-phenotypic prediction of A. thaliana samples8 is at the broad end of the spectrum, since it involved predicting hundreds of real-valued phenotypes, such as root length, flowering time, seed dormancy, etc. These papers show that the GenGI approach I propose can be a reality, and it already provides promising preliminary results. They contain in nuce the core aspects that I will develop to a full-fledged computational framework during the 5 years of my current CPJ position at IGMM.
cases versus controls binary discrimination, while in the broad sense the goal is to predict multiple phenotypes for each sample. The papers on end-to-end GI that I have recently published 8,9,10,11 are the embryos from which I will expand this project. In particular, our Crohn’s disease (CD) case-control predictors8,9,10,11 belonged to the narrow sense GenGI, while Galiana, the multi-phenotypic prediction of A. thaliana samples8 is at the broad end of the spectrum, since it involved predicting hundreds of real-valued phenotypes, such as root length, flowering time, seed dormancy, etc. These papers show that the GenGI approach I propose can be a reality, and it already provides promising preliminary results. They contain in nuce the core aspects that I will develop to a full-fledged computational framework during the 5 years of my current CPJ position at IGMM.
References
1- https://www.nature.com/articles/nrg.2016.86 ;
2- https://pmc.ncbi.nlm.nih.gov/articles/PMC2493042/ ;
3- onlinelibrary.wiley.com/doi/full/10.1002/humu.23280;
4- https://pubmed.ncbi.nlm.nih.gov/19859063/ ;
5- https://pmc.ncbi.nlm.nih.gov/articles/PMC4143101/ ;
6- https://www.nature.com/articles/s41586-021-03819-2 ;
7- https://www.nature.com/articles/nature08250 ;
8- https://pubmed.ncbi.nlm.nih.gov/34792168/ ;
9- academic.oup.com/nargab/article/2/1/lqaa011/5742219 ;
10- https://doi.org/10.1186/s13059-023-03064-y ;
11- https://www.nature.com/articles/s41598-023-46887-2
Selected Publications
[1] N. Verplaetse, A. Passemiers, A. Arany, Y Moreau, D. Raimondi. “Large sample size and nonlinear sparse models outline epistatic effects in inflammatory bowel disease”. Genome Biology 24 (1), 224 (2023)
[2] D. Raimondi, M. Corso, P. Fariselli, Y. Moreau, From genotype to phenotype in Arabidopsis thaliana: in-silico genome interpretation predicts 288 phenotypes from sequencing data, Nucleic Acids Research, 2021; gkab1099, https://doi.org/10.1093/nar/gkab1099 2022
[3] D. Raimondi, J. Simm, A. Arany, Y. Moreau, A novel method for data fusion over entity-relation graphs and its application to protein–protein interaction prediction, Bioinformatics, Volume 37, Issue 16, 15 August 2021, Pages 2275–2281, https://doi.org/10.1093/bioinformatics/btab092
List of publications
*Raimondi, Daniele, Antoine Passemiers, Nora Verplaetse, Massimiliano Corso, Ángel Ferrero-Serrano, Nelson Nazzicari, Filippo Biscarini, Piero Fariselli, and Yves Moreau. "Biologically meaningful genome interpretation models to address data underdetermination for the leaf and seed ionome prediction in Arabidopsis thaliana." Scientific reports 14, no. 1 (2024): 13188.
*Gavalda-Garcia, J., Bickel, D., Roca-Martinez, J., Raimondi, D., Orlando, G., & Vranken, W. (2024). Data-driven probabilistic definition of the low energy conformational states of protein residues. NAR genomics and bioinformatics, 6(3), lqae082.
* Passemiers, A., Tuveri, S., Sudhakaran, D., Jatsenko, T., Laga, T., Punie, K., ... & Vermeesch, J. R. (2024). MetDecode: methylation-based deconvolution of cell-free DNA for noninvasive multi-cancer typing. Bioinformatics, 40(9), btae522.
*Nourisa, J., Passemiers, A., Shakeri, F., Omidi, M., Helmholz, H., Raimondi, D., ... & Zeller-Plumhoff, B. (2024). Gene regulatory network analysis identifies MYL1, MDH2, GLS, and TRIM28 as the principal proteins in the response of mesenchymal stem cells to Mg2+ ions. Computational and structural biotechnology journal, 23, 1773-1785.
*Passemiers, A., Folco, P., Raimondi, D., Birolo, G., Moreau, Y., & Fariselli, P. (2024). A quantitative benchmark of neural network feature selection methods for detecting nonlinear signals. Scientific Reports, 14(1), 31180.
* D. Raimondi, H. Chizari, N. Verplaetse, B.S. Löscher, A. Franke, Y. Moreau. “Genome interpretation in a federated learning context allows the multi-center exome-based risk prediction of Crohn’s disease patients” Scientific Reports 13 (1), 19449 (2023)
* N. Verplaetse, A. Passemiers, A. Arany, Y Moreau, D. Raimondi. “Large sample size and nonlinear sparse models outline epistatic effects in inflammatory bowel disease”. Genome Biology 24 (1), 224 (2023)
* Mazzone, Eugenio, Yves Moreau, Piero Fariselli, and Daniele Raimondi. "Nonlinear data fusion over Entity–Relation graphs for Drug–Target Interaction prediction." Bioinformatics 39, no. 6 (2023): tad348.
* Raimondi, Daniele, Francesco Codicè, Gabriele Orlando, Joost Schymkowitz, Frederic Rousseau, and Yves Moreau. "HPMPdb: A machine learning-ready database of protein molecular phenotypes associated to human missense variants." Current Research in Structural Biology 4 (2022): 167-174.
* Orlando, Gabriele, Daniele Raimondi, Francesco Codice, Francesco Tabaro, and Wim Vranken. Prediction of disordered regions in proteins with recurrent neural networks and protein dynamics. Journal of Molecular Biology 434, no. 12 (2022): 167579.
* Passemiers, Antoine, Yves Moreau, and Daniele Raimondi. Fast and accurate inference of gene regulatory networks through robust precision matrix estimation. Bioinformatics 38, no. 10 (2022): 2802-2809.
* G. Orlando, D. Raimondi, R. Duran-Romana, Y. Moreau, J. Schymkowitz, F. Rousseau, PyUUL: an interface between biological structures and deep learning algorithms, Nature Communications, 13 (1), 961 (2022)
* D. Raimondi, M. Corso, P. Fariselli, Y. Moreau, From genotype to phenotype in Arabidopsis thaliana: in-silico genome interpretation predicts 288 phenotypes from sequencing data, Nucleic Acids Research, 2021; gkab1099, https://doi.org/10.1093/nar/gkab1099 2022
* D. Raimondi, A. Passemiers, P. Fariselli, and Y. Moreau. Current cancer driver variant predictors learn to recognize driver genes instead of functional variants. BMC Biology, 19(1), 2021.
* L.P. Kagami, G. Orlando, D. Raimondi, F. Ancien, B. Dixit, J. Gavaldá-Garciá, P. Ramasamy, J. Roca-Martı́ nez, K. Tzavella, and W. Vranken. B2btools: Online predictions for protein biophysical features and their conservation. Nucleic Acids Research, 49(W1):W52–W59, 2021.* M. Necci, et al., Critical assessment of protein intrinsic disorder prediction. Nature Methods, 18(5):472–481, 2021.
* D. Raimondi, J. Simm, A. Arany, Y. Moreau, A novel method for data fusion over entity-relation graphs and its application to protein–protein interaction prediction, Bioinformatics, Volume 37, Issue 16, 15 August 2021, Pages 2275–2281, https://doi.org/10.1093/bioinformatics/btab092
* D. Raimondi, G. Orlando, E. Michiels, D. Pakravan, A. Bratek-Skicki, L. Van Den Bosch, Y. Moreau, F. Rousseau, J. Schymkowitz, In silico prediction of in vitro protein liquid–liquid phase separation experiments outcomes with multi-head neural attention, Bioinformatics, Volume 37, Issue 20, 15 October 2021, Pages 3473–3479, https://doi.org/10.1093/bioinformatics/btab350
* L.M. Peeters, et al., Covid-19 in people with multiple sclerosis: A global data sharing initiative. Multiple Sclerosis Journal, 26(10):1157–1162, 2020.
* G. Orlando, D. Raimondi, L. P. Kagami, W. F Vranken, ShiftCrypt: a web server to understand and biophysically align proteins through their NMR chemical shift values, Nucleic Acids Research, Volume 48, Issue W1, 02 July 2020, Pages W36–W40, https://doi.org/10.1093/nar/gkaa391
* G. Orlando, A. Silva, S. MacEdo-Ribeiro, D. Raimondi, and W. Vranken. Accurate prediction of protein beta-aggregation with generalized statistical potentials. Bioinformatics, 36(7):2076–2081, 2020.
* G. Buroni, Y.-A. Le Borgne, G. Bontempi, D. Raimondi, and K. Determe. On-board unit big data: Short-term traffic forecasting in urban transportation networks. 2020 IEEE 7th International Conference on Data Science and Advanced Analytics (DSAA), 2020, pp. 569-578, doi:10.1109/DSAA49011.2020.00072. 2020
* D. Raimondi, J. Simm, A. Arany, P. Fariselli, I. Cleynen, Y. Moreau, An interpretable low-complexity machine learning framework for robust exome-based in-silico diagnosis of Crohn’s disease patients, NAR Genomics and Bioinformatics, Volume 2, Issue 1, March 2020, lqaa011, https://doi.org/10.1093/nargab/lqaa011
* D. Raimondi, G. Orlando, P. Fariselli, and Y. Moreau. Insight into the protein solubility driving forces with neural attention. PLoS Computational Biology, 16(4), 2020. * D. Raimondi, G. Orlando, W.F. Vranken, and Y. Moreau. Exploring the limitations of biophysical propensity scales coupled with machine learning for protein sequence analysis. Scientific Reports, 9(1), 2019.
* G. Orlando, D. Raimondi, F. Tabaro, F. Codicè, Y. Moreau, and W.F. Vranken. Computational identification of prion-like rna-binding proteins that form liquid phase-separated condensates. Bioinformatics, 35(22):4617–4623, 2019.
* G. Orlando, D. Raimondi, and W. F. Vranken. Auto-encoding nmr chemical shifts from their native vector space to a residue-level biophysical index. Nature Communications, 10(1), 2019.
* D. Raimondi, G. Orlando, Y. Moreau, and W.F. Vranken. Ultra-fast global homology detection with discrete cosine transform and dynamic time warping. Bioinformatics, 34(18):3118–3125, 2018.
* D. Raimondi, G. Orlando, F. Tabaro, T. Lenaerts, M. Rooman, Y. Moreau, and W.F. Vranken. Large-scale in-silico statistical mutagenesis analysis sheds light on the deleteriousness landscape of the human proteome. Scientific Reports, 8(1), 2018.
* D. Raimondi, I. Tanyalcin, J.S.D. Fertè, A. Gazzo, G. Orlando, T. Lenaerts, M. Rooman, and W. Vranken. Deogen2: Prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Research, 45(W1):W201–W206, 2017.
* G. Orlando, D. Raimondi, T. Khan, T. Lenaerts, and W.F. Vranken. Svm-dependent pairwise hmm: An application to protein pairwise alignments. Bioinformatics, 33(24):3902–3908, 2017.
* A. Gazzo, D. Raimondi, D. Daneels, Y. Moreau, G. Smits, S. Van Dooren, and T. Lenaerts. Understanding mutational effects in digenic diseases. Nucleic Acids Research, 45(15), 2017.
* D. Raimondi, G. Orlando, R. Pancsa, T. Khan, and W.F. Vranken. Exploring the sequence-based prediction of folding initiation sites in proteins. Scientific Reports, 7(1), 2017.
* D. Raimondi, G. Orlando, J. Messens, and W.F. Vranken. Investigating the molecular mechanisms behind uncharacterized cysteine losses from prediction of their oxidation state. Human Mutation, 38(1):86–94, 2017.
* D. Raimondi, A.M. Gazzo, M. Rooman, T. Lenaerts, and W.F. Vranken. Multilevel biological characterization of exomic variants at the protein level significantly improves the identification of their deleterious effects. Bioinformatics, 32(12):1797–1804, 2016.
* R. Pancsa, D. Raimondi, E. Cilia, and W. Vranken. Early folding events, local interactions, and conservation of protein backbone rigidity. Biophysical Journal, 110(3):572–583, 2016.
* G. Orlando, D. Raimondi, and W.F. Vranken. Observation selection bias in contact prediction and its implications for structural bioinformatics. Scientific Reports, 6, 2016.
* D. Raimondi, G. Orlando, and W.F. Vranken. An evolutionary view on disulfide bond connectivities prediction using phylogenetic trees and a simple cysteine mutation model. PLoS ONE, 10(7), 2015.
* D. Raimondi, G. Orlando, and W.F. Vranken. Clustering-based model of cysteine co-evolution improves disulfide bond connectivity prediction and reduces homologous sequence requirements. Bioinformatics, 31(8):1219–1225, 2015.
* M.J. Skwark, D. Raimondi, M. Michel, and A. Elofsson. Improved contact predictions using the recognition of protein like contact patterns. PloS Computational Biology, 10(11), 2014.
Pre-prints:
* Sudhakaran, Dhanya, Stefania Tuveri, Antoine Passemiers, Tatjana Jatsenko, Tina Laga, Kevin Punie, Sabine Tejpar et al. "MetDecode: methylation-based deconvolution of cell-free DNA for non-invasive multi-cancer typing." medRxiv (2023): 2023-12.
* J Gavalda-Garcia, D Bickel, J Roca-Martinez, D Raimondi, G Orlando, Wim Vranken. “Data-driven probabilistic definition of the low energy conformational states of protein residues” bioRxiv, 2023.07. 24.550386 (2023)
* Passemiers, Antoine, Pietro Folco, Daniele Raimondi, Giovanni Birolo, Yves Moreau, and Piero Fariselli. "How good Neural Networks interpretation methods really are? A quantitative benchmark." arXiv preprint arXiv:2304.02383 (2023).
* Edward De Brouwer, Daniele Raimondi, Yves Moreau. Modeling the COVID-19 outbreaks and the effectiveness of the containment measures adopted across countries. doi: https://doi.org/10.1101/2020.04.02.20046375
* Gabriele Orlando, Daniele Raimondi, Francesco Codice, Francesco Tabaro, Wim Vranken Prediction of disordered regions in proteins with recurrent Neural Networks and protein dynamics doi: https://doi.org/10.1101/2020.05.25.115253
* Edward De Brouwer, Daniele Raimondi, Yves Moreau. Can herd immunity be achieved without breaking ICUs? doi: https://doi.org/10.1101/2020.05.26.20113746
Editorials:
* Raimondi, Daniele, Gabriele Orlando, Nora Verplaetse, Piero Fariselli, and Yves Moreau. "Towards genome interpretation: Computational methods to model the genotype-phenotype relationship." Frontiers in Bioinformatics 2 (2022): 1098941.



