Our research aims to investigate the intricate interactions between RNA, TFs and processing factors and how they contribute to cell fate decisions.
Integrating transcription and mRNA processing networks into cell fate decision
-intRNAnet-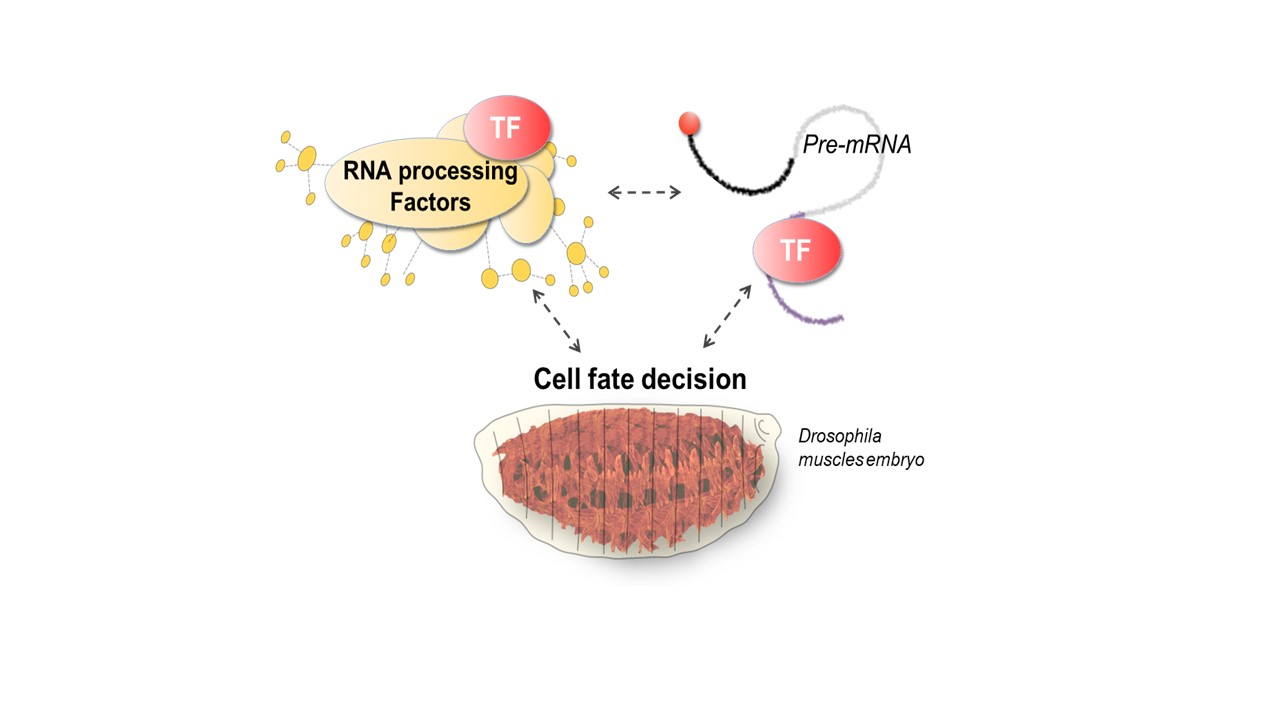
OVERVIEW
Gene expression, the key to cell diversity across all organisms, is governed by transcription factors (TFs) orchestrating intricate gene regulatory networks with precise spatial and developmental timings. Yet, significant challenges persist in elucidating the complexity and dynamics of these networks, as well as in connecting gene expression to cell fate decisions.
Beyond the conventional view that TFs act solely on cis-regulatory DNA elements (promoters, enhancers), we and others have demonstrated that many TFs can bind RNA and modulate mRNA fates. By doing so, TFs regulate a unique layer of molecular diversity through the production of various mRNA isoforms, thus shaping specific proteomes and contributing to cell diversity. Despite its central role in gene networks, the contribution of TF-RNA regulatory functions to cell fate decisions remains largely unexplored. Our research team aims to unravel these unconventional TF-RNA functions and their impact on cell fate changes, both on developmental processes and on dysregulated states observed in diseases.
OUR QUESTIONS
Pre-mRNA processing is a conserved process covering events such as 5’capping, splicing and 3’polyadenylation. It relies on the dynamic assembly of molecular machines, notably the spliceosome, to produce mature mRNAs in precise cell contexts and ensures the production of various mRNA isoforms with great plasticity. Our research goal is to investigate the intricate interactions between RNA, TFs and processing factors, and how they contribute to cell fate decisions.
Specifically, we aim to understand:
1- TF functions on pre-mRNA fate: How do TFs integrate transcription and splicing functions? Do TFs regulate other processing layers?
2- The functional specificity: What drives TF-RNA interaction specificity, and how do these networks coordinate cell fate changes?
3- The conservation of RNA regulation: Are TF-RNA functions universal? Which TFs bind RNA, and how?
OUR APPROACHES
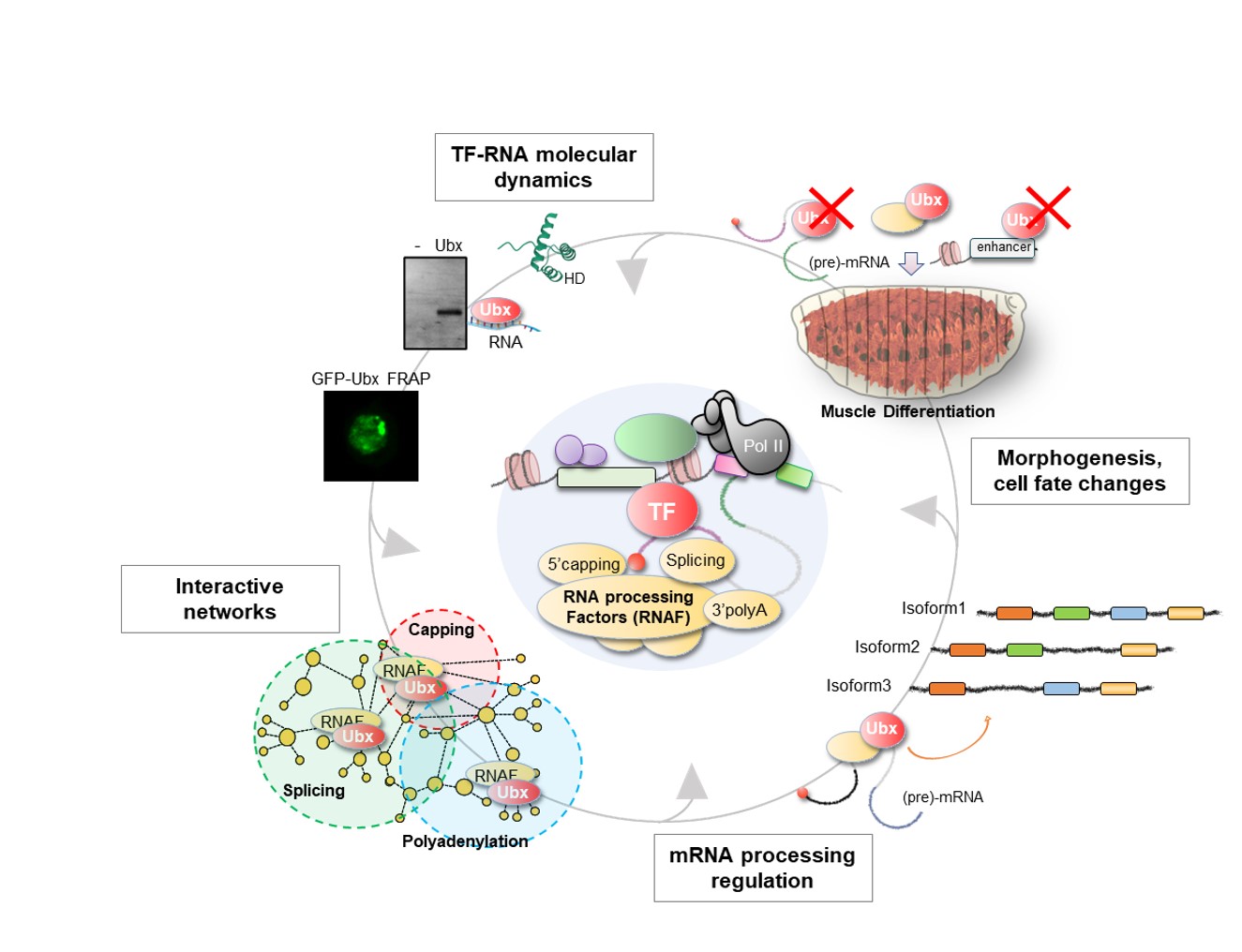
To explore these questions, we focus on the homeodomain (HD) TFs and primarily on the Hox TFs, which are prominent players of cell identity and tissue patterning in animals (see the figure below exemplified with the Drosophila Hox TF Ultrabithorax, our work showed that Ubx regulates alternative splicing). We investigate their role at multiple scales: in vitro, in cell culture and in vivo in Drosophila embryonic muscles, for which the identity and function are regulated at the co-transcriptional and post-transcriptional levels.
Our research is rooted in molecular developmental biology and aims to bridge structural biology and in silico modelling through stimulating collaborations for studying TF-RNA networks from molecules to tissue development. Specifically, we employ biochemistry (RNA-DNA-protein interaction assays), transcriptomics (RNA-seq, RIP-seq), proteomics (BioID, split-BioID coupled with mass spectrometry) and imaging methods (RNA, protein, tissue patterning) combined with in vivo Drosophila genetics (mutants, nanobodies toolkit) for integrative molecular and functional analyses of TF-RNA regulatory functions.
OUR TEAM

Recruitment announcements will be available soon for a staff engineer (IE) on the CNRS platform and Master students.
We will be happy to hear from you if you are interested in the lab’s research!
Please contact Julie and include your CV and a brief outline of why you are interested in joining the lab.
Selection of publications :
-Synergistic DNA and RNA binding of the Hox transcription factor Ultrabithorax coordinates splicing and shapes in vivo homeotic functions. Blanco C, Xiang W, Boumpas P, Scorcelletti M, Hermon AS, Wong J, Merabet S, Carnesecchi J*. 2024 doi: https://doi.org/10.1101/2024.09.10.612310. *corresponding author
-Integrating transcription and splicing into cell fate: Transcription factors on the block. Boumpas P, Merabet S, Carnesecchi J*. Wiley Interdiscip Rev RNA. 2023 Mar;14(2):e1752. PMID: 35899407. *corresponding author
-The Hox transcription factor Ultrabithorax binds RNA and regulates co-transcriptional splicing through an interplay with RNA polymerase II. Carnesecchi J*, Boumpas P, van Nierop Y Sanchez P, Domsch K, Pinto HD, Borges Pinto P, Lohmann I. Nucleic Acids Res. 2022 Jan 25;50(2):763-783. PMID: 34931250. *corresponding author
-Multi-level and lineage-specific interactomes of the Hox transcription factor Ubx contribute to its functional specificity. Carnesecchi J, Sigismondo G, Domsch K, Baader CEP, Rafiee MR, Krijgsveld J, Lohmann I. Nat Commun. 2020 Mar 13;11(1):1388. PMID: 32170121.
List of all the publication :
-Sleiman HN, Carnesecchi J, Jia Y, Delolme F, Gilquin L, Gouet P, Merabet S. An improved nanobody-based approach to capture and visualize specific interaction networks of binary protein complexes in living cells. bioRxiv. doi: https://doi.org/10.1101/2024.09.12.612471
-Hox dosage and morphological diversification during development and evolution. Merabet S, Carnesecchi J. Semin Cell Dev Biol. 2024 Jan-Feb;152-153:70-75. PMID: 36481343.
-Integrating transcription and splicing into cell fate: Transcription factors on the block. Boumpas P, Merabet S, Carnesecchi J. Wiley Interdiscip Rev RNA. 2023 Mar;14(2):e1752. PMID: 35899407.
-Specificity of the Hox member Deformed is determined by transcription factor levels and binding site affinities. Pinto PB, Domsch K, Gao X, Wölk M, Carnesecchi J, Lohmann I. Nat Commun. 2022 Aug 26;13(1):5037. PMID: 36028502
-The Hox transcription factor Ultrabithorax binds RNA and regulates co-transcriptional splicing through an interplay with RNA polymerase II. Carnesecchi J, Boumpas P, van Nierop Y Sanchez P, Domsch K, Pinto HD, Borges Pinto P, Lohmann I. Nucleic Acids Res. 2022 Jan 25;50(2):763-783. PMID: 34931250.
-Role of a versatile peptide motif controlling Hox nuclear export and autophagy in the Drosophila fat body. Duffraisse M, Paul R, Carnesecchi J, Hudry B, Banreti A, Reboulet J, Ajuria L, Lohmann I, Merabet S. J Cell Sci. 2020 Sep 23;133(18). PMID: 32878938.
-Closing the gap: a roadmap to single-cell regulatory genomics. Carnesecchi J, Lohmann I. Mol Syst Biol. 2020 May;16(5):e9497. PMID: 32430985.
- Multi-level and lineage-specific interactomes of the Hox transcription factor Ubx contribute to its functional specificity. Carnesecchi J, Sigismondo G, Domsch K, Baader CEP, Rafiee MR, Krijgsveld J, Lohmann I. Nat Commun. 2020 Mar 13;11(1):1388. PMID: 32170121.
-The Hox transcription factor Ubx stabilizes lineage commitment by suppressing cellular plasticity in Drosophila. Domsch K, Carnesecchi J, Disela V, Friedrich J, Trost N, Ermakova O, Polychronidou M, Lohmann I. Elife. 2019 May 3;8. PMID: 31050646.
-LSD1-ERRα complex requires NRF1 to positively regulate transcription and cell invasion. Zhang L, Carnesecchi J, Cerutti C, Tribollet V, Périan S, Forcet C, Wong J, Vanacker JM. Sci Rep. 2018 Jul 3;8(1):10041. PMID: 29968728.
-Hox transcription factors: an overview of multi-step regulators of gene expression. Carnesecchi J, Pinto PB, Lohmann I. Int J Dev Biol. 2018;62(11-12):723-732. PMID: 30604842.
-ERRα protein is stabilized by LSD1 in a demethylation-independent manner. Carnesecchi J, Cerutti C, Vanacker JM, Forcet C. PLoS One. 2017;12(11):e0188871. PMID: 29190800.
-ERRα induces H3K9 demethylation by LSD1 to promote cell invasion. Carnesecchi J, Forcet C, Zhang L, Tribollet V, Barenton B, Boudra R, Cerutti C, Billas IM, Sérandour AA, Carroll JS, Beaudoin C, Vanacker JM. Proc Natl Acad Sci U S A. 2017 Apr 11;114(15):3909-3914. PMID: 28348226.
-Estrogen-Related Receptors and the control of bone cell fate. Carnesecchi J, Vanacker JM. Mol Cell Endocrinol. 2016 Sep 5;432:37-43. PMID: 26206717.
-Estrogens induce rapid cytoskeleton re-organization in human dermal fibroblasts via the non-classical receptor GPR30. Carnesecchi J, Malbouyres M, de Mets R, Balland M, Beauchef G, Vié K, Chamot C, Lionnet C, Ruggiero F, Vanacker JM. PLoS One. 2015;10(3):e0120672. PMID: 25781607.
-Estrogen-related receptor α decreases RHOA stability to induce orientated cell migration. Sailland J, Tribollet V, Forcet C, Billon C, Barenton B, Carnesecchi J, Bachmann A, Gauthier KC, Yu S, Giguère V, Chan FL, Vanacker JM. Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15108-13. PMID: 25288732.
-The estrogen-related receptors and the adipocyte. Carnesecchi J, Vanacker JM. Horm Mol Biol Clin Investig. 2013 Aug;14(3):107-12. PMID: 25436725.
-Repression of osteoblast maturation by ERRα accounts for bone loss induced by estrogen deficiency. Gallet M, Saïdi S, Haÿ E, Photsavang J, Marty C, Sailland J, Carnesecchi J, Tribollet V, Barenton B, Forcet C, Birling MC, Sorg T, Chassande O, Cohen-Solal M, Vanacker JM. PLoS One. 2013;8(1):e54837. PMID: 23359549.



