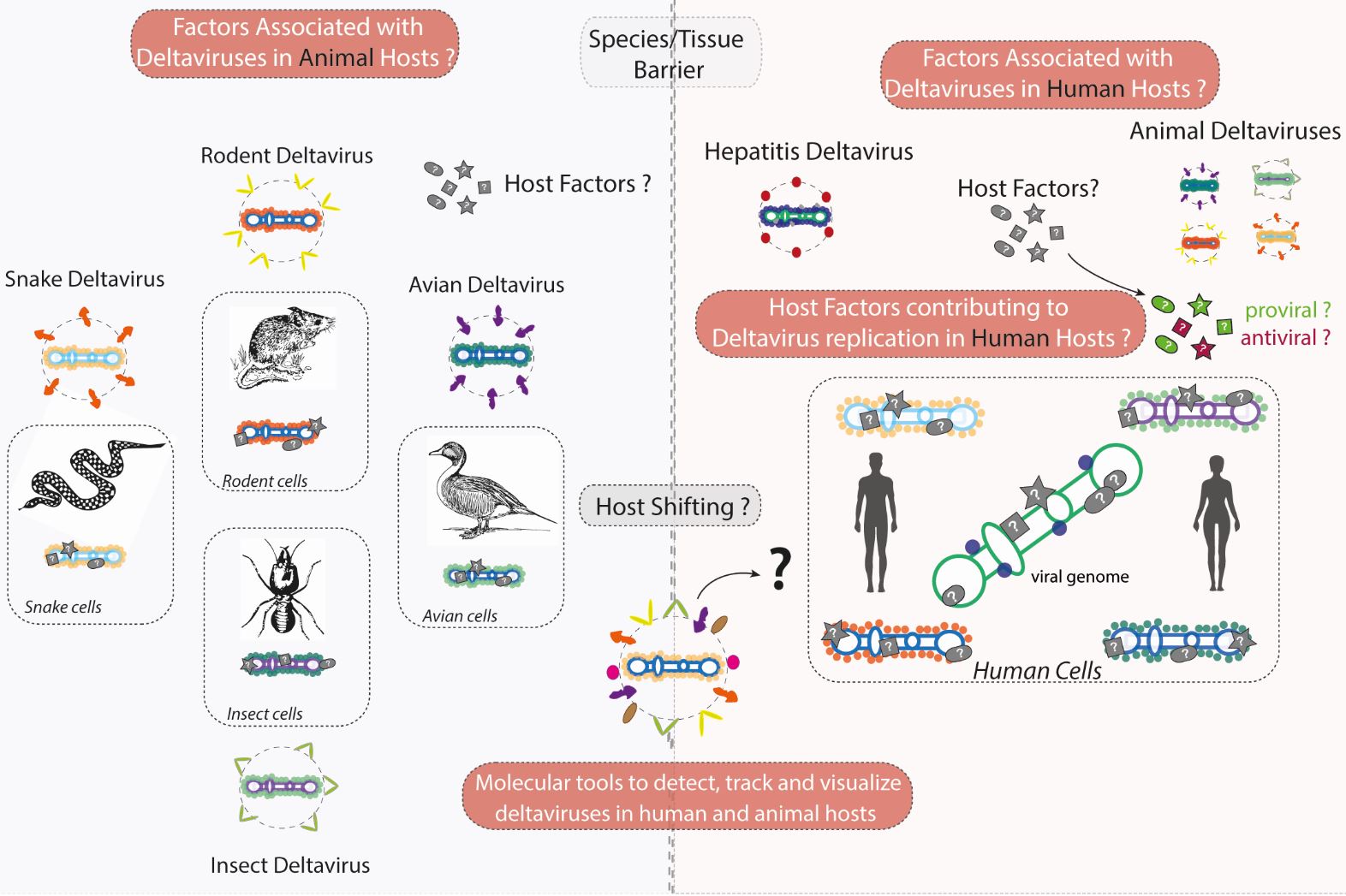
As obligate intracellular parasites, RNA viruses are normally restricted to their animal host, having established over long evolutionary periods intimate interactions with cellular host factors. In fact, RNA viruses rely heavily on host factors to complete their lifecycle during translation, transcription or replication of viral genomes. Viral interactions with cellular host factors largely determine the evolutionary success of a virus and the disease it may cause. Deltaviruses being the smallest known viruses (by genome size) to infect animals, are the “quintessential” viruses, depending almost exclusively on host factors to complete their replication cycle. Using deltaviruses as simple RNA virus models, the aim of our group is to identify and characterize host factors essential for replication of RNA viruses in both human and animal hosts.
For more than 40 years the only known member of deltaviruses was Hepatitis D Virus, a virus with no available antiviral cure, infecting 15 to 20 million individuals worldwide and leading to a very severe form of viral hepatitis and liver cancer. We have known for few years now that HDV-like viruses (referred to as deltaviruses) are present in a variety of animal vectors and reservoirs including bats, rats, snakes, birds, and insects. Importantly, we now also know that deltaviruses are not restricted to the liver, as previously thought, and can replicate in a variety of animal tissues, possessing a previously unrecognized host shifting capacity. Our group is using and building new molecular tools to study and understand deltavirus infection in both human and animal hosts.
1- We are building new molecular tools to detect, track and visualize deltavirus genomes in human, rodent, snake, avian and insect cell culture models. These include cloning of novel deltavirus replicon and reporter systems coupled to novel microscopy methods to detect host and viral proteins and RNAs (e.g. nanobodies, SunTAG, RNA FISH).
2- We employ nucleic acid and protein purification methods coupled to proteomics (e.g. IP-MS, ChIRP-MS) to systematically evaluate Delta RNA-Host protein interactions in human, rodent, snake, avian and insect cell culture models. Our goal is to determine the protein and RNA-interactomes of complexes associated to animal deltaviruses, identifying universal, conserved and/or specific host factors that play roles in deltavirus replication and infection across the animal kingdom.
3- We use genetic screening approaches (e.g. CRIPSR knock-out, CRISPR activation) to identify, characterize and target pro-viral and antiviral factors controlling replication of HDV and animal deltaviruses in human cells. Our goal is to better understand the replication of these viruses in human cells and to generate a list of potential host targets for the development of new antiviral strategies.
Legend : 3D reconstitution of a persistently replicating Huh7.5.1-HDV nucleus.
Confocal Z-stack images from the persistently replicating nucleus. Viral RNA is in red and viral proteins in green.
Legend : 3D reconstitution of a persistently replicating NIH-3T3-RDeV nucleus.
Confocal Z-stacks images from the persistently replicating nucleus. Viral RNA is in red and viral proteins in green.
Legend : 3D reconstitution of a persistently replicating I/1Ki-SDeV nucleus.
Confocal Z-stacks images from the persistently replicating nucleus. Viral RNA is in red and viral proteins in green.


Zoe DENIS (PhD Student)
In LAB :
Identify and characterize host factors involved in:
- The restriction of Deltaviruses (ISGs) through an ISG screening
- The replication and the persistence of Deltaviruses through a genome-wide CRISPR-Cas9 screen
Outside LAB :
Likes Climbing (serious climber) and traveling
Pierre KHALFI (Post-Doc)
In LAB :
- Setting up tools and readouts to study the replication of deltaviruses (e.g.stable deltaviruses expressing cell lines, mouse model, smFISH, Northern Blot.)
- Comprehensive identification of deltavirus RNA-binding proteins by mass spectrometry (ChIRP-MS). I am now using these tools to determine which of the host factors I have identified can control
Outside LAB :
Likes Scuba diving, cycling and cocktails
Joe MCKELLAR (Post-Doc)
In LAB :
- Resolving the structure of Deltavirus RNPs in the context of the cell or in viral particles by cryo-EM.
- Developing new techniques to visualize Deltavirus infections in cells.
- Understanding the rules that govern Deltavirus assembly with glycoproteins of helper viruses.
Outside LAB :
I am an amateur home cook and enjoy participating in competitive Magic The Gathering tournaments.
Roni SLEIMAN (PhD Student)
In LAB:
- Resolve Deltavirus particle and RNP structure by Cryo-EM techniques
- Development of new molecular tools to detect , track and visualize Delta RNPs.
Outside LAB:
In his free time, seeks one of two things; Either peace and quiet, or a blast with friends over food and cocktails!
Louison BOIS (Master 2 Intern)
In LAB:
Investigates the replication of circular RNA viruses Deltaviruses and Ambiviruses in the two divergent yeast species, Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Outside LAB :
Enjoys running, climbing and going out with friends
Leila BLAIR (Lab Manager)
In LAB:
- Implement and develop molecular tools for the study of Deltaviruses.
- Ensure efficiency in laboratory manipulations through resource management and the creation and maintenance of organizational systems.
Outside LAB :
When I'm not in the lab I enjoy going for walks in nature spaces, volunteering, cooking/baking, and making art, like watercolor, pastels or collage.
Valérie COURGNAUD (CRHC/ Scientist)
In LAB :
-Focuses on the impact of interactions between viruses and their hosts, mainly on retroviruses, HDVs and delta-like viruses in animals.
I'm interested in viral evolution and cross-species transmission. Using genetic screens, my projects currently focus on identifying genes involved in resistance to viral infection,
with a particular interest in interferon-regulated proteins and on deciphering the mechanisms involved in these processes.
Outside lab :
Reading, swimming, and lot of others things…..



