In a work published in Genome Research, Marie-Odile Baudement and its colleagues (team « Genome organization and epigenetic control » led by Thierry Forné), have developed the HRS-seq, a new straightforward genome-wide approach whereby they isolated and sequenced genomic regions associated with nuclear bodies and other membrane-less nuclear organelles which are made insoluble through high-salt treatments (HRS – High-salt Recovered Sequences). Their works demonstrated that, in mouse embryonic stem cells (ESC), these regions essentially correspond to the most highly expressed genes, and to cis-regulatory sequences like super-enhancers, that belong to the active A chromosomal compartment. They include both cell type-specific genes, such as pluripotency genes in ESC, and housekeeping genes associated with nuclear bodies, such as histone and snRNA genes that are central components of Histone Locus Bodies and Cajal bodies. These results, which are in agreement with the recently proposed “phase separation model” for transcription control (Hnisz et al., 2017, Cell 169,13-23), suggest that nuclear bodies might play a central role in organizing the active chromosomal compartment in mammals.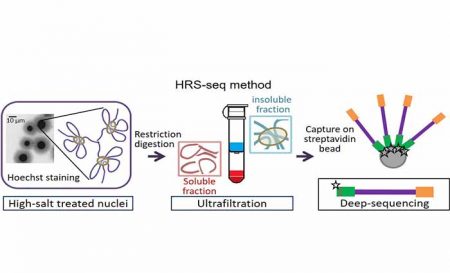
Baudement M-O., Cournac A., Court F., Seveno M., Parrinello H., Reynes C., Sabatier R., Bouschet T., Yi Z., Sallis S., Tancelin M., Rebouissou C., Cathala G., Lesne A., Mozziconacci* J., Journot* L. and Forné* T. « High-salt Recovered Sequences are associated with the active chromosomal compartment and with large ribonucleoprotein complexes including nuclear bodies ». Genome Research, in press.
Published in Advance, October 4, 2018, doi: 10.1101/gr.237073.118.