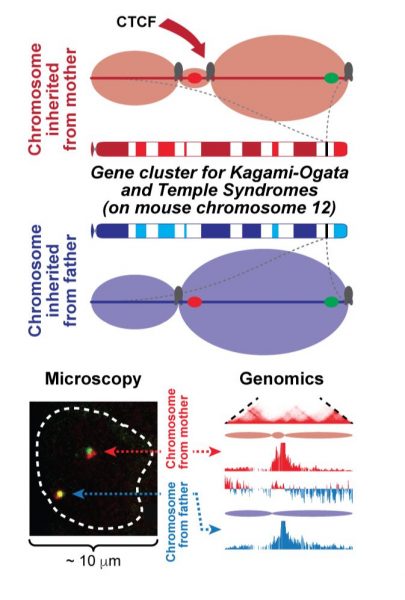
In their study published in Genome Biology, David Llères and his co-workers (Genomic Imprinting and Development group headed by Robert Feil) explains how the structure of DNA controls the genes that are responsible for several of complex imprinting disorders.
The genes that are involved in four of these human imprinting disorders (Beckwith-Wiedemann, Silver-Russell, Kagami-Ogata and Temple Syndromes) are organized in two large gene clusters, where the DNA methylation imprint is present on the chromosome from the father. Using mouse cells as a model, Robert Feil’s tam and their collaborators (Daan Noordermeer‘s lab, Institute for Integrative Biology of the Cell, Gif-sur-Yvette, France) found that the CTCF factor, a well-known protein that helps folding chromosomes, binds to additional positions in both clusters when inherited from the mother (where the imprint is absent). By combining high-resolution genomics and microscopy approaches, the scientists were then able to demonstrate that this additional CTCF binding creates a new level of physical DNA domain structure that is unique to the maternal chromosome and that this difference in folding controls the opposing patterns of gene activity between the parental chromosomes.
To know more:
CTCF modulates allele-specific sub-TAD organisation and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains.
David Llères, Benoît Moindrot, Rakesh Pathak, Vincent Piras, Mélody Matelot, Benoît Pignard, Alice Marchand, Mallory Poncelet, Aurélien Perrin, Virgile Tellier, Robert Feil* and Daan Noordermeer*. Genome Biology, 2019, 20:272. DOI: https://doi.org/10.1186/s13059-019-1896-8