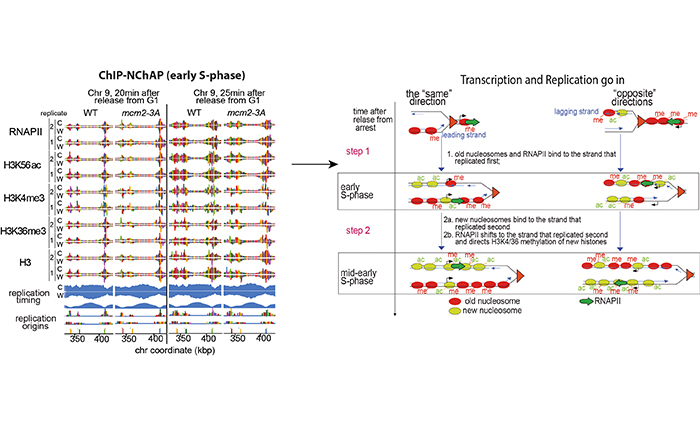
We developed ChIP-NChAP (Nascent Chromatin Avidin Pulldown)-a method to monitor budding yeast replication, transcription and chromatin maturation dynamics on each daughter genome in parallel. We find a difference in the timing of lagging and leading strand replication on the order of minutes at most yeast genes. We propose a model in which the majority of old histones and RNAPII bind to the gene copy that replicated first, while newly synthesized nucleosomes are assembled on the copy that replicated second. RNAPII enrichment then shifts to the sister copy that replicated second. The order of replication is largely determined by genic orientation: if transcription and replication are co-directional the leading strand replicates first; if they are counter-directional the lagging strand replicates first. We now propose that active transcription states are inherited simultaneously and independently of their underlying chromatin states through the recycling of the transcription machinery and old histones, respectively. Transcription thus actively contributes to the reestablishment of the active chromatin state.
Ziane, R., Camasses, A. & Radman-Livaja, M. The asymmetric distribution of RNA polymerase II and nucleosomes on replicated daughter genomes is caused by differences in replication timing between the lagging and the leading strand. Genome Res, doi:10.1101/gr.275387.121 (2022).