Asymmetric cell division generates cell diversity across all kingdoms of life. We use the budding yeast Saccharomyces cerevisiae as a model system to understand the mechanisms and functions of asymmetric cell division.
Yeast cells are also a very good model of cellular ageing. Damages such as protein aggregates accumulate in the mother cell. How protein aggregates are asymmetrically inherited to allow rejuvenation of daughter cells is still poorly understood. Beyond identifying the mechanisms of asymmetric cell division, we follow the idea that ageing allows cells to keep memories of their past adaptations to cope better with future stress.
1.Prion-like proteins can store cellular memory.
We discovered a new type of epigenetic memory when yeast cells encounter a deceptive mating attempt (Caudron & Barral, Cell, 2013). When two haploid cells of opposite sexual type meet, they communicate through sexual pheromone secretion, and both arrest their cell cycle in the G1 phase. Mating partners then grow towards each other by forming a shmoo. When they reach each other, they fuse and become a diploid cell. However, life is not always that simple. When cells are exposed to pheromone only, without a mating partner in reach, they first shmoo and then can escape the pheromone-induced arrest, re-enter the cell cycle and produce daughter cells. Mother cells maintain this pheromone refractory state for the remainder of their lifespan. In contrast, daughter cells are born naïve and respond to pheromone (Figure 1).
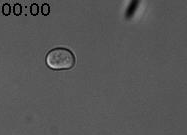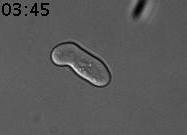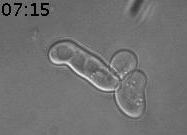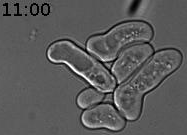
Figure 1. Cells were exposed to alpha-factor (7nM). The mother cell is at first shmooing and then only produces daughter cells, which shmoo. Time is in hours and minutes.
The pheromone refractory state depends on the inactivation of Whi3. Whi3 inhibits translation of mRNA encoding Cln3, the G1 cyclin required for the G1 to S phase transition. Whi3 contains prion-like domains that promote an inactivating conformational change upon pheromone treatment. We termed this type of protein mnemon. A mnemon is a protein that change of fold and super-assembly can encode a cellular memory. It is specifically induced by a stimulus and inherited asymmetrically during cell division (Figure 2).
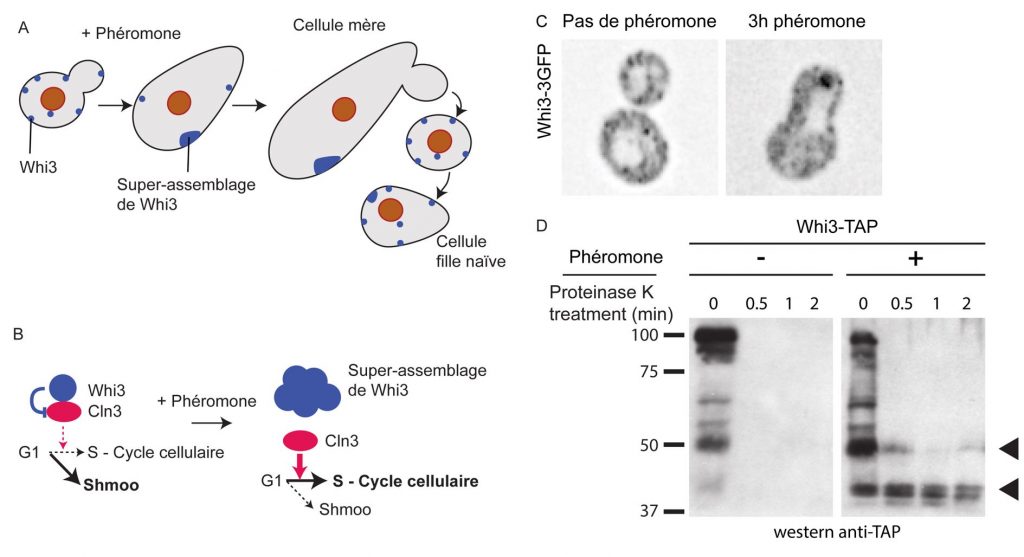
Figure 2. Schematic of the deceptive mating memory in budding yeast. A. A budding yeast cell exposed to pheromone shmoos. Whi3 super-assembles slowly, allowing the cell to escape the pheromone induced cell cycle arrest and produce a bud. The bud is born naive, free of Whi3 super-assembly and respond to pheromone. B. Upon pheromone treatment, Whi3 is inhibited through its super-assembly. The translation of Cln3 mRNA is no more repressed by Whi3, and Cln3 can commit cells to the S-phase. C. Whi3-3GFP localization in untreated and pheromone-exposed cells. Note the super-assembly of Whi3 after pheromone treatment. D. Biochemical characterization of Whi3’s conformation change. Whi3 becomes partially resistant to proteinase K treatment after cells were exposed to pheromone.
In contrast, prions are symmetrically inherited during cell division. We have identified that Whi3 association with the membranes of the endoplasmic reticulum (ER) promotes its retention in the mother cell. This is due to the compartmentalization of the ER membranes by lateral diffusion membranes at the bud neck, the limit between the mother cell and the future daughter cell. Disruption of the diffusion barrier leads to the transformation of Whi3 from a mnemon to a symmetrically inherited prion (Lau et al., Current Biology, 2022, Figure 3). Proteins with prion-like domains are very common in most living organisms and many of these molecules cause proteopathies such as neurodegenerative diseases or Creutzfeldt-Jakob disease. We test the idea that many proteins behave as mnemons, and we aim at understanding their biology in physiological and pathological contexts.
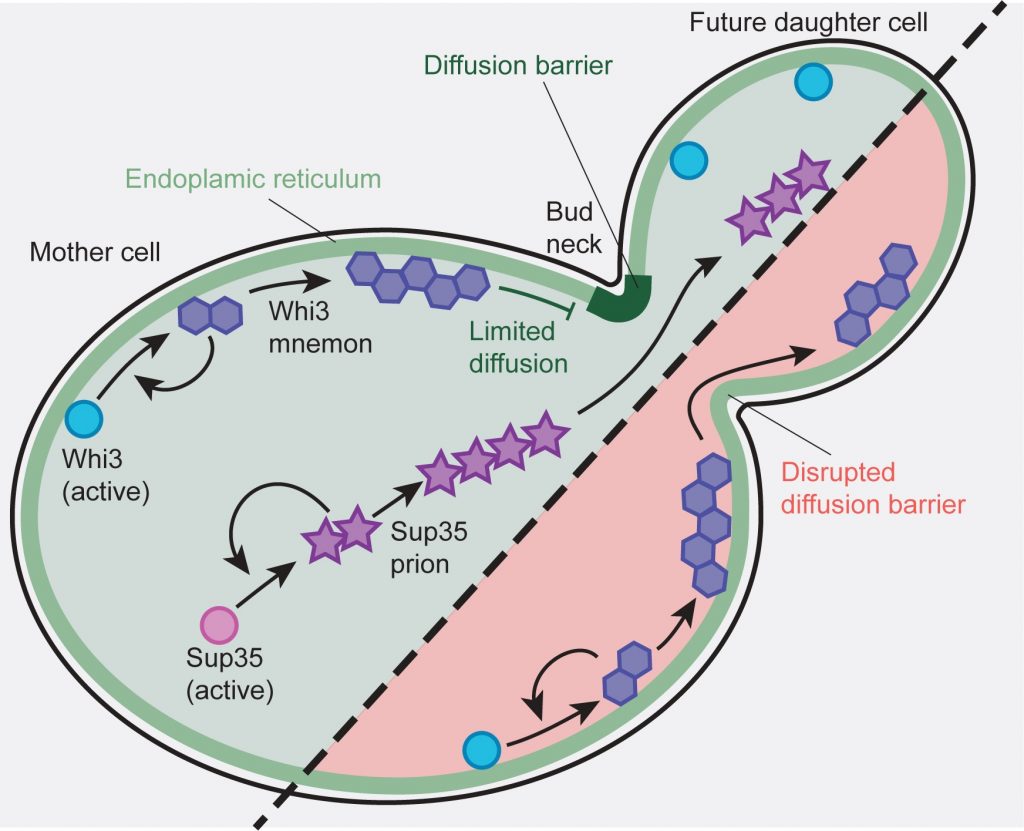
Figure 3. The Whi3 mnemon associates with endoplasmic reticulum membranes. A diffusion barrier at the bud neck restricts Whi3’s propagation to daughter cells. The prion form of Sup35, not associated with membranes is free to diffuse to daughter cells. When the diffusion barrier is genetically weakened, the Whi3 mnemon can propagate to daughter cells as a prion.
2.Fate of prion-like proteins during ageing.
Ageing impacts most living organisms with an array of physiological functions that decline. Cellular ageing is accompanied by the accumulation of protein in an aggregated form. In budding yeast, a single inclusion containing aggregated proteins is nearly always inherited by the mother cell during cell division (Saarikangas et al., Current Biology, 2017, Figure 4). We aim at understanding how protein aggregates are inherited during cell division as well as identifying which properties of old cell induce the formation of these aggregates. We particularly focus on prion-like proteins, because the mnemon Whi3 does aggregate in old cells (Schlissel et al., Science, 2017, Figure 4). To achieve this, we use microfluidic chips to follow the full life span of single yeast cells.

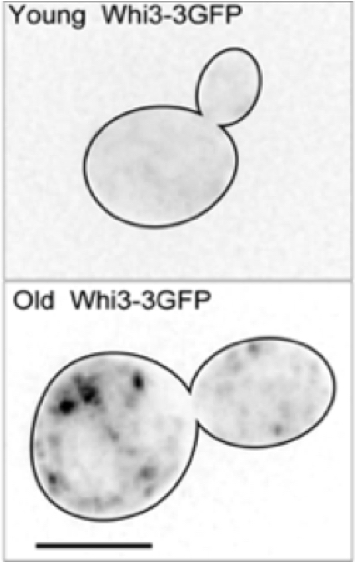
Figure 4. The left panel shows a movie of an old yeast cell expressing Hsp104-GFP. The age-induced protein deposit is seen as a dot that remains in the mother cell. The right panel shows a young yeast cell (top) expressing the mnemon Whi3-GFP. The signal is diffuse, while in an old yeast cell (bottom) it forms aggregates.
3.Identification of molecules that can delay aging.
In an increasingly aging population, we hope that the detrimental aspects of aging could be delayed, to increase healthspan. In this context, we have identified that the molecule tripentadecanoin can delay aging of yeast cells through inhibition of age-induced protein aggregates formation (Oamen et al., BioRXiv, 2021, Figure 5). We are working actively on identifying how tripentadecanoin works at the molecular level and try to identify other compounds that could promote healthspan.
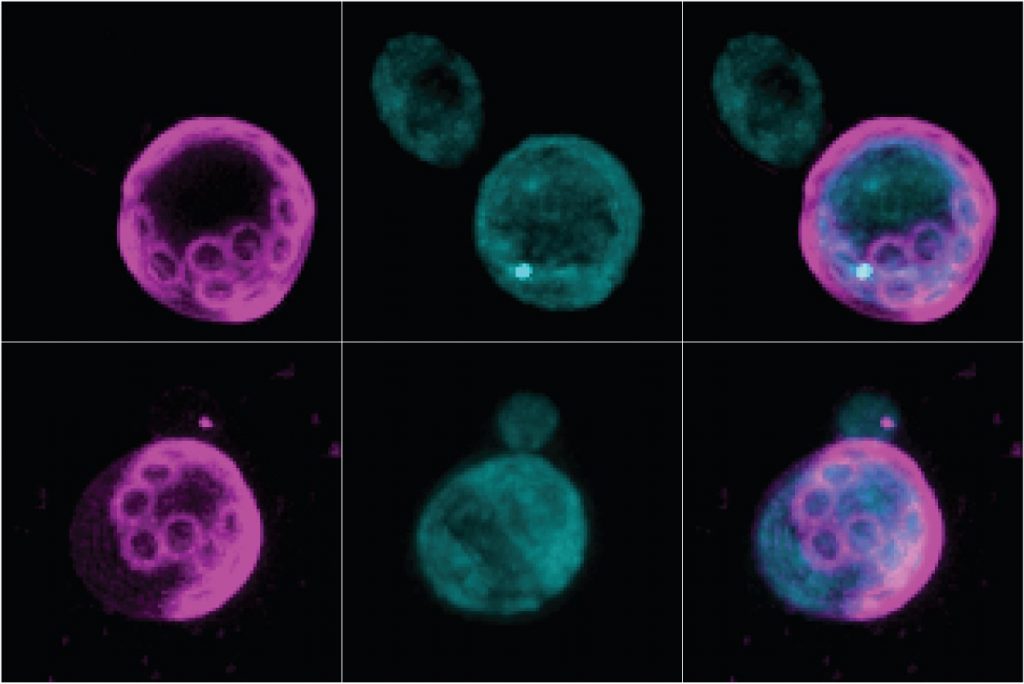
Figure 5. Bud scars (the rings, left) show the age of old cells. The protein Hsp104-GFP forms an age-induced aggregate (dot, middle upper panel) in untreated cells. Cells treated with tripentadecanoin (bottom) do not display this aggregate and live longer.
Selection of publications :
Whi3 mnemon association with endoplasmic reticulum membranes confines the memory of deceptive courtship to the yeast mother cell. Lau, Y., Oamen, H.P., Grogg, M., Parfenova, I., Saarikangas, J., Hannay, R., Nichols, R.A., Hilvert, D., Barral, Y., and Caudron, F. (2022). Current Biology.
Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells. Schlissel, G., Krzyzanowski, M.K., Caudron, F., Barral, Y., and Rine, J. (2017). Science 355, 1184–1187.
A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Caudron, F., and Barral, Y. (2013). Cell 155, 1244–1257.
List of all the publications :
1. A rare natural lipid induces neuroglobin expression to prevent amyloid oligomers toxicity and retinal neurodegeneration. Oamen, H.P., Romero, N.R., Knuckles, P., Saarikangas, J., Dong, Y., and Caudron, F. (2021). bioRxiv, 20. https://doi.org/10.1101/2021.06.23.449608
2. Whi3 mnemon association with endoplasmic reticulum membranes confines the memory of deceptive courtship to the yeast mother cell. Lau, Y., Oamen, H.P., Grogg, M., Parfenova, I., Saarikangas, J., Hannay, R., Nichols, R.A., Hilvert, D., Barral, Y., and Caudron, F. (2022). Current Biology.
3. Mnemons and the memorization of past signaling events. Reichert, P., and Caudron, F. (2021). Curr Opin Cell Biol 69, 127–135. PMID: 33618243
4. Prion-like proteins as epigenetic devices of stress adaptation. Oamen, H.P., Lau, Y., and Caudron, F. (2020). Experimental Cell Research 396, 112262. PMID: 32896568
5. Protein Phase Separation during Stress Adaptation and Cellular Memory. Lau, Y., Oamen, H.P., and Caudron, F. (2020). Cells 9, 1302. PMID: 32456195
6. Meeting report – shining light on septins. Caudron, F., and Yadav, S. (2018). Journal of Cell Science 131. PMID: 29360625
7. A Droplet to Sense Sugar Drops. Barral, Y., and Caudron, F. (2017). Molecular Cell 68, 1017–1019. PMID: 29272701
8. Spatial regulation of coalesced protein assemblies: Lessons from yeast to diseases. Saarikangas, J., and Caudron, F. (2017). Prion 259, 1–12. PMID: 28574744
9. Protein aggregation triggers a declining libido in elder yeasts that still have a lust for life. Caudron, F. (2017). Microbial Cell 4, 200–202. PMID: 28660204
10. Compartmentalization of ER-Bound Chaperone Confines Protein Deposit Formation to the Aging Yeast Cell. Saarikangas, J., Caudron, F., Prasad, R., Moreno, D.F., Bolognesi, A., Aldea, M., and Barral, Y. (2017). Current Biology 27, 773–783. PMID: 28262489
11. Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells. Schlissel, G., Krzyzanowski, M.K., Caudron, F., Barral, Y., and Rine, J. (2017). Science 355, 1184–1187. PMID: 28302853
12. A role for the yeast CLIP170 ortholog, the plus-end-tracking protein Bik1, and the Rho1 GTPase in Snc1 trafficking. Boscheron, C., Caudron, F., Loeillet, S., Peloso, C., Mugnier, M., Kurzawa, L., Nicolas, A., Denarier, E., Aubry, L., and Andrieux, A. (2016). Journal of Cell Science 129, 3332–3341. PMID: 27466378
13. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Clay, L., Caudron, F., Denoth-Lippuner, A., Boettcher, B., Buvelot Frei, S., Snapp, E.L., and Barral, Y. (2014). eLife 3, e01883. PMID: 24843009
14. [Sex and memory: what yeast can teach us]. Caudron, F., and Barral, Y. (2014). Médecine Sciences 30, 348–349. PMID: 24801024
15. Mnemons: encoding memory by protein super-assembly. Caudron, F., and , Barral, Y. (2014). Microbial Cell 1, 100–102. PMID: 28357228
16. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Caudron, F., and Barral, Y. (2013). Cell 155, 1244–1257. PMID: 24315096
17. Bud building by septin patch hole punching. Caudron, F., and Barral, Y. (2013). Developmental Cell 26, 115–116. PMID: 23906060
18. Mutation of Ser172 in yeast β tubulin induces defects in microtubule dynamics and cell division. Caudron, F., Denarier, E., Thibout-Quintana, J.-C., Brocard, J., Andrieux, A., and Fourest-Lieuvin, A. (2010). PloS One 5, e13553. PMID: 21042413
19. Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit. Manzoni, R., Montani, F., Visintin, C., Caudron, F., Ciliberto, A., and Visintin, R. (2010). The Journal of Cell Biology 190, 209–222. PMID: 20660629
20. Septins and the lateral compartmentalization of eukaryotic membranes. Caudron, F., and Barral, Y. (2009). Developmental Cell 16, 493–506. PMID: 19386259
21. [A diffusion barrier allows budding yeast rejuvenation]. Caudron, F. (2009). Médecine Sciences 25, 217–218. PMID: 19361377
22. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. Caudron, F., Andrieux, A., Job, D., and Boscheron, C. (2008). Journal of cell science 121, 1506–1513. PMID: 18411245




