Inflammation is a normal reaction of the immune system in response to infections or tissue damage. Inflammation is a component of the innate immune response that is elicited by a broad range of triggers, comprises a broad range of mechanisms and signaling pathways, all of which converging towards the production of soluble mediators of inflammation (cytokines and chemokines). Beneficial inflammatory responses are generally initiated locally, at the site of infection or injury, and constitute a central signaling step towards the activation of adaptive immune responses. Once the infection cleared or the wound healed, the inflammatory response is resolved, in order to limit the activation of the immune system over time.
When inflammatory responses are unresolved and persist in time, they are pervasive and may lead to systemic damage, affecting tissues heterogeneously. As a consequence, chronic inflammation is associated with a vast range etiologically distinct human pathologies, including cancer, auto-inflammatory and auto-immune disorders, but also metabolic diseases or neurodegenerative disorders and other non-communicable diseases. As such, chronic inflammation associated human pathologies weigh heavily on both socio-economic and healthcare systems, calling for innovative therapeutic routes.
The goal of the Molecular Basis of Inflammation laboratory is to provide a comprehensive view of the pathways governing the activation, maintenance and resolution of inflammatory responses. Furthermore, we aim to uncover the moelcualar mechanisms governing tissue-specific inflammatory responses. Our current research interests revolve around the study of inflammatory responses triggered by pathological nucleic acid species, that include cytosolic dsDNAs, ssDNAs and RNA:DNA hybrids. These moieties can arise in the cytosol following genotoxic or mitochondrial stress or following pathogen infection. Although they are all known to trigger inflammatory processes, it is also well established that they promote cell type-specific signatures, that may in turn promote differential tissue responses. To decipher how deregulation of inflammatory processes promote pathologies, we combine a broad array of approaches, ranging from mechanistic studies in vitro, to phenotyping in vivo. Combining several scales of study should allow charting the impact of pathological nucleic acids on cellular pathways and assessment of their impact at the whole organism level, potentially identifying key signalling nodes that can be acted upon to hinder pathological inflammation. In a nutshell:
• At the molecular level: we identify and characterize new pathways involved in triggering inflammatory pathologies,
• At the cellular level: we characterize how these pathways are regulated depending on cellular identity,
• At the tissue level: we study how cellular heterogeneity within tissues dictates the outcome of inflammatory response and impacts homeostasis
• At the organism level: we study inflammatory responses and the consequences of their pathological activation on integrated in vivo models.
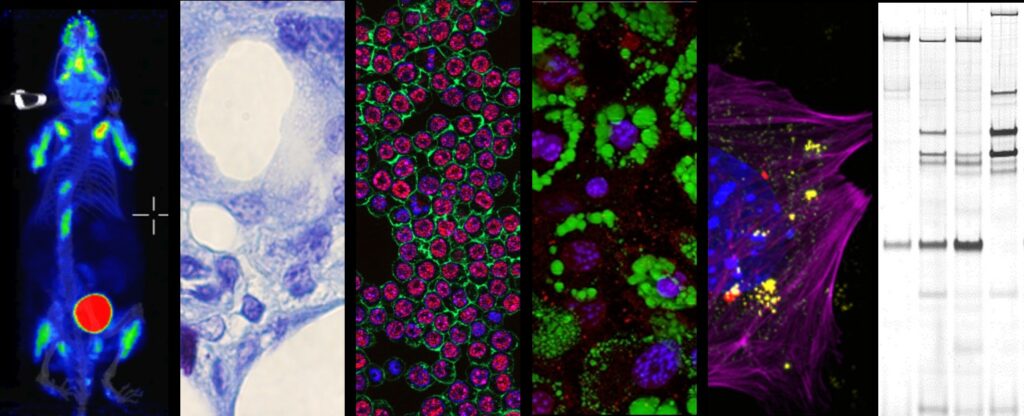
Current interests of the Molecular Basis of Inflammation Team revolve around the theme of nucleic acid immunity. Pathological nucleic acids susceptible of triggering uninflammatory responses include cytosolic dsDNA, ssDNAs and DNA:RNA hybrids. Those may arise in the cytosol of cells following pathogen infection, genotoxic or mitochondrial stress and/or aberrant endogenous retroelement activity. Several detection routes for cytosolic nucleic acid species have been described in the past decade, although the molecular mechanisms underlying their cell type-specific activation and coordination remain poorly described. Additionally, for each given nucleic acid trigger, the ensuing inflammatory response and the associated physiopathological consequences, are governed by parameters that remain to be uncovered.
The major team’s projects aim to describe the molecular mechanisms involved in the activation of nucleic acid immunity, study their evolutionary conservation, and unravel how they feed pathological inflammation and tissue-specific alterations.
Projects developed by the team include:
• Detection and regulation of cytosolic DNA:RNA hybrids in tumorigenesis
• Novel cytosolic nucleic acids detection routes
• The crosstalk between inflammatory and metabolic responses
• Chromatin control of inflammatory responses
• ssDNA detection and regulation in chronic and acute inflammatory responses
• Cyclic dinucleotide in the control of inflammatory and physiological responses
• Harnessing co-regulators of nucleic acid detection pathways for boosting antitumoral responses
1.Detection and regulation of cytosolic DNA:RNA hybrids in tumorigenesis
We have shown that the reverse transcription (RT) activity associated with endogenous retroelements generates immunogenic cytosolic nucleic acids. These nucleic acids are recognized via the cGAS-STING pathway, central to the detection of cytosolic nucleic acids. Activation of this signaling pathway feeds chronic inflammation associated with the Fanconi Anemia cancer predisposition syndrome (Brégnard et al. EBioMed, 2016).
We next focused on identifying pathways regulating RT-generated immunogenic nucleic acids - in particular DNA:RNA hybrids. We have identified the Lysyl-tRNA synthetase (LysRS), a key player in protein translation of, as involved in the regulation of inflammation associated with the cytosolic DNA:RNA hybrids (Guerra et al. Science Adv, 2020).
Currently, we are studying the impact of LysRS on tumorigenesis, notably in the context of pancreatic adenocarcinoma (PDAC).
2.Novel cytosolic nucleic acids detection routes

Genotoxic stress is a complex phenomenon engaging numerous molecular interactions and post-translational modifications, induced by genetic instability and DNA replication stress. Genotoxic stress response proteins are direct regulators of the innate immune response via (i) their ability to recognize non-canonical nucleic acid structures, including endogenous DNAs present in the cytosol, and (ii) regulation of DNA repair. Our recent work has established that the DNA-PK holoenzyme, involved in the repair of double strand breaks by non-homologous end-joining, plays a major role in boosting the activation of the canonical cGAS-STING pathway, which is involved in the detection of cytosolic dsDNAs (Taffoni et al 2022). Using a range of in vivo and in vitro approaches, we showed that DNA-PK and cGAS cooperate to drive cancer-associated inflammatory responses, ultimately regulating the breadth of anti-tumor immune responses.
Because several tumors present with mutations in DNA-PK or cGAS, we now wish to investigate whether the cooperation of these two pathways could be harnessed to boost anti-tumoral responses, or to the contrary prevent deleterious, cancer-promoting inflammatory responses.
3.The crosstalk between inflammatory and metabolic responses

Chronic inflammation can be fueled by high fat diets that promote concerted alterations of inflammatory and metabolic pathways. We recently identified the STING protein, central to the production of type I Interferons in the presence of cytosolic nucleic acids, as a central regulator of polyunsaturated fatty acid (PUFA) metabolism (Vila et al. Cell Metabolism 2022). We are exploring the communication between these inflammatory and metabolic pathways, at the cellular, tissue and whole-body levels, to reveal their impact on the maintenance of homeostasis and in chronic inflammatory pathologies.
To this aim:
(i) We interrogate the role of potential imbalances in PUFA desaturation in pathologies presenting with chronic STING-dependent inflammation
(ii) We investigate whether metabolic regulation is the primordial function of STING.
(iii) We question how type I IFN responses are achieved in metabolic cells.
4.Chromatin control of inflammatory responses
Although most immunogenic nucleic acids are detected in the cytosol, the proteins mediating this recognition are, to a large extent, nuclear and associated with the chromatin when inactive. This for example the case of the cGAS protein, central to the detection of nucleic acids in the cytosol. Our current work shows that the cytosolic presence of immunogenic nucleic acids leads to the mobilization of numerous chromatin proteins, beyond already described receptors. We study the consequences of cytosolic mobilization of nuclear proteins and their impact on the regulation of inflammatory responses.
5.ssDNA detection and regulation in chronic and acute inflammatory responses
Physiologically, ssDNAs occur in cells upon replication stress and resolution of DNA damage. In cancer cells and certain immunopathologies, ssDNAs seem to accumulate and trigger inflammatory signaling. In addition, ssDNA can be pathogen-associated molecular patterns (PAMP) that arise during viral infection. In contrast to double-stranded DNA (dsDNA), the mechanisms of and requirements for ssDNA sensing are poorly defined. We wish to increase the understanding of ssDNA sensing and to better define the sensors, pathways and modulators involved.
6.Cyclic dinucleotide in the control of inflammatory and physiological responses
The cyclic AMP-GMP (cGAMP) second messenger is synthesized by the cGAMP synthase (cGAS) upon detection of cytosolic nucleic acids, including dsDNAs. cGAMP is the major activator of the adaptor protein STING and therefore plays a crucial role in the activation of type I Interferon and inflammatory responses. The production of cGAMP is a tightly regulated process for which, for example, we have shown that the DNA-PK DNA repair complex is necessary (Taffoni et al, EMBOJ 2022). Beyond this function of cGAMP in STING activation, there is growing evidence the implication of cyclic dinucleotides in numerous cellular functions, such as the regulation of polyunsaturated fatty acid metabolism (Vila et al Cell Metabolism 2022). In addition, dinucleotides of bacterial origin can elicit inflammatory responses in host cells upon infection. Altogether these evidences suggest that cyclic dinucleotides such as cGAMP are major players in the regulation of homeostasis.
We currently study the molecular mechanisms involved in the regulation of the production, propagation and degradation of cyclic dinucleotides.
7.Harnessing co-regulators of nucleic acid detection pathways for boosting antitumoral responses
The STING adaptor protein is a target of growing interest for the reactivation of anti-tumor immunity. We have identified several STING partners and screened for inhibitors and co-activators, amongst which druggable target have been validated. We wish to validate the efficiency of identified therapeutic combinations for their ability to boost anti-tumour responses in preclinical models of breast cancer, melanoma and pancreatic adenocarcinoma.
Selected Publications
Towards a novel approach to stimulate anti-tumoral immunity in glioblastoma. Taffoni C, Laguette N. Med Sci (Paris). 2023 Nov;39(11):813-815. doi: 10.1051/medsci/2023149. PMID: 38018919
Editorial: Broadening our view on nucleic acid sensing: novel sensors, signaling pathways, and involvement in non-infectious diseases. Chauveau L, Laguette N, Sampaio NG. Front Immunol. 2023 Aug 11;14:1266732. doi: 10.3389/fimmu.2023.1266732. eCollection 2023. PMID: 37638050
Multiplexed targeted analysis of polyunsaturated fatty acids and oxylipins using liquid chromatography-tandem mass spectrometry. Turtoi E, Jeudy J, Valette G, Enjalbal C, Vila IK, Laguette N, Turtoi A. STAR Protoc. 2023 Aug 17;4(3):102226. doi: 10.1016/j.xpro.2023.102226. PMID: 37597187
The unexpected role of the STING protein in lipid metabolism. Vila IK, Laguette N. C R Biol. 2023 Apr 18;346:29-33. doi: 10.5802/crbiol.110. PMID: 37254782
Dietary restriction induces a sexually dimorphic type I interferon response in mice with gene-environment interactions. Harney DJ, Cielesh M, Roberts GE, Vila IK, Viengkhou B, Hofer MJ, Laguette N, Larance M. Cell Rep. 2023 Jun 27;42(6):112559. doi: 10.1016/j.celrep.2023.112559. May 26. PMID: 37243595
Harnessing the cooperation between DNA-PK and cGAS in cancer therapies: The cooperation between DNA-PK and cGAS shapes tumour immunogenicity. Taffoni C, Schüssler M, Vila IK, Laguette N. Bioessays. 2023 Jul;45(7):e2300045. doi: 10.1002/bies.202300045. May 5. PMID: 37147791
DNA damage repair kinase DNA-PK and cGAS synergize to induce cancer-related inflammation in glioblastoma. Taffoni C, Marines J, Chamma H, Guha S, Saccas M, Bouzid A, Valadao AC, Maghe C, Jardine J, Park MK, Polak K, De Martino M, Vanpouille-Box C, Del Rio M, Gongora C, Gavard J, Bidère N, Song MS, Pineau D, Hugnot JP, Kissa K, Fontenille L, Blanchet FP, Vila IK, Laguette N. EMBO J. 2022 Dec 27:e111961. doi: 10.15252/embj.2022111961. Online ahead of print. PMID: 36574362
Regulation of innate immunity by Nrf2. van der Horst D, Carter-Timofte ME, van Grevenynghe J, Laguette N, Dinkova-Kostova AT, Olagnier D. Curr Opin Immunol. 2022 Oct;78:102247. doi: 10.1016/j.coi.2022.102247. Epub 2022 Sep 26. PMID: 36174411
Alternative pathways driven by STING: From innate immunity to lipid metabolism. Vila IK, Guha S, Kalucka J, Olagnier D, Laguette N. Cytokine Growth Factor Rev. 2022 Dec;68:54-68. doi: 10.1016/j.cytogfr.2022.08.006. Epub 2022 Sep 1.PMID: 36085258 Review.
Activation of STING in the pancreatic tumor microenvironment: A novel therapeutic opportunity. Chamma H, Vila IK, Taffoni C, Turtoi A, Laguette N. Cancer Lett. 2022 Jul 10;538:215694. doi: 10.1016/j.canlet.2022.215694. Epub 2022 Apr 27. PMID: 35489447 Review.
Protocol to induce and assess cGAS-STING pathway activation in vitro. Chamma H, Guha S, Laguette N, Vila IK. STAR Protoc. 2022 May 14;3(2):101384. doi: 10.1016/j.xpro.2022.101384. eCollection 2022 Jun 17. PMID: 35600929
STING orchestrates the crosstalk between polyunsaturated fatty acid metabolism and inflammatory responses. Vila IK, Chamma H, Steer A, Saccas M, Taffoni C, Turtoi E, Reinert LS, Hussain S, Marines J, Jin L, Bonnefont X, Hubert M, Schwartz O, Paludan SR, Van Simaeys G, Doumont G, Sobhian B, Vlachakis D, Turtoi A, Laguette N. Cell Metab. 2022 Jan 4;34(1):125-139.e8. doi: 10.1016/j.cmet.2021.12.007. PMID: 34986331
Editorial: Nucleic Acid-Associated Inflammation. Laguette N, Langevin C, Olagnier D, Torraca V, Vanpouille-Box C, Verrier ER. Front Immunol. 2021 Oct 27;12:791580. doi: 10.3389/fimmu.2021.791580. eCollection 2021. PMID: 34777401
Nucleic Acid Immunity and DNA Damage Response: New Friends and Old Foes. Taffoni C, Steer A, Marines J, Chamma H, Vila IK, Laguette N. Front Immunol. 2021 Apr 26;12:660560. doi: 10.3389/fimmu.2021.660560. eCollection 2021. PMID: 33981307 Review.
Animal Models for the Study of Nucleic Acid Immunity: Novel Tools and New Perspectives. Vila IK, Fretaud M, Vlachakis D, Laguette N, Langevin C. J Mol Biol. 2020 Sep 18;432(20):5529-5543. doi: 10.1016/j.jmb.2020.08.016. Epub 2020 Aug 26. PMID: 32860771. Review.
Lysyl-tRNA synthetase produces diadenosine tetraphosphate to curb STING-dependent inflammation. Guerra J, Valadao AL, Vlachakis D, Polak K, Vila IK, Taffoni C, Prabakaran T, Marriott AS, Kaczmarek R, Houel A, Auzemery B, Déjardin S, Boudinot P, Nawrot B, Jones NJ, Paludan SR, Kossida S, Langevin C, Laguette N. Sci Adv. 2020 May 22;6(21):eaax3333. doi: 10.1126/sciadv.aax3333. eCollection 2020 May. PMID: 32494729
V1b vasopressin receptor trafficking and signaling: Role of arrestins, G proteins and Src kinase. Perkovska S, Méjean C, Ayoub MA, Li J, Hemery F, Corbani M, Laguette N, Ventura MA, Orcel H, Durroux T, Mouillac B, Mendre C. Traffic. 2018 Jan;19(1):58-82. doi: 10.1111/tra.12535. PMID: 29044966
Upregulated LINE-1 Activity in the Fanconi Anemia Cancer Susceptibility Syndrome Leads to Spontaneous Pro-inflammatory Cytokine Production. Brégnard C, Guerra J, Déjardin S, Passalacqua F, Benkirane M, Laguette N. EBioMedicine. 2016 Jun;8:184-194. doi: 10.1016/j.ebiom.2016.05.005. Epub 2016 May 6. PMID: 27428429
Shaping of the host cell by viral accessory proteins. Laguette N, Benkirane M. Front Microbiol. 2015 Feb 23;6:142. doi: 10.3389/fmicb.2015.00142. eCollection 2015. PMID: 25755653
DNA damage repair machinery and HIV escape from innate immune sensing. Brégnard C, Benkirane M, Laguette N. Front Microbiol. 2014 Apr 22;5:176. doi: 10.3389/fmicb.2014.00176. eCollection 2014. PMID: 24795708
Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Laguette N, Brégnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. Cell. 2014 Jan 16;156(1-2):134-45. doi: 10.1016/j.cell.2013.12.011. Epub 2014 Jan 9. PMID: 24412650
Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Cell Rep. 2013 Apr 25;3(4):1036-43. doi: 10.1016/j.celrep.2013.03.017. Epub 2013 Apr 17. PMID: 23602554
SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. Retrovirology. 2012 Oct 23;9:87. doi: 10.1186/1742-4690-9-87. PMID: 23092122
Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Brandariz-Nuñez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. Retrovirology. 2012 Jun 12;9:49. doi: 10.1186/1742-4690-9-49. PMID: 22691373
SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. Nat Immunol. 2012 Feb 12;13(3):223-228. doi: 10.1038/ni.2236. PMID: 22327569
Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Münch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. Cell Host Microbe. 2012 Feb 16;11(2):205-17. doi: 10.1016/j.chom.2012.01.007. Epub 2012 Feb 1. PMID: 22305291
How SAMHD1 changes our view of viral restriction. Laguette N, Benkirane M. Trends Immunol. 2012 Jan;33(1):26-33. doi: 10.1016/j.it.2011.11.002. Epub 2011 Dec 15. PMID: 22177690
SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. Nature. 2011 May 25;474(7353):654-7. doi: 10.1038/nature10117. PMID: 21613998



