All living cells rely on the establishment of particular gene expression programs to sustain basic cellular functions, acquire specific identities and respond to a variety of extracellular and intracellular stimuli. The first and likely most regulated step in gene expression is the transcription of genomic DNA by RNA polymerases, which generate distinct repertoires of RNA transcripts (transcriptomes) depending on the cell type and environmental conditions. However, while transcription is absolutely essential, it is also a source of potential conflicts with other processes co-occurring in the genome, such as DNA replication and the repair of DNA damage. If unresolved, these conflicts can compromise genome integrity.
We seek to elucidate the pathways that shape transcriptomes and the mechanisms that regulate the conflicts caused by transcription machineries. We also aim at understanding how perturbations in these processes are involved in human diseases. We employ a variety of approaches ranging from high-resolution genomics, molecular genetics, and biochemistry and we use different eukaryotic model systems such as budding yeast and various types of human cells, including motor neurons.
Our research is structured into 4 different axes :
AXIS 1:
Study of the mechanism of transcription termination for non-coding and protein-coding genes
(PIs: D. Libri & O. Porrua)
Transcription is not limited to regions annotated for producing known functional RNAs, but occurs virtually everywhere in the genome, a phenomenon conserved from bacteria to humans that is named pervasive or hidden transcription (see our reviews Villa & Porrua, 2022; and Jensen et al, 2013).
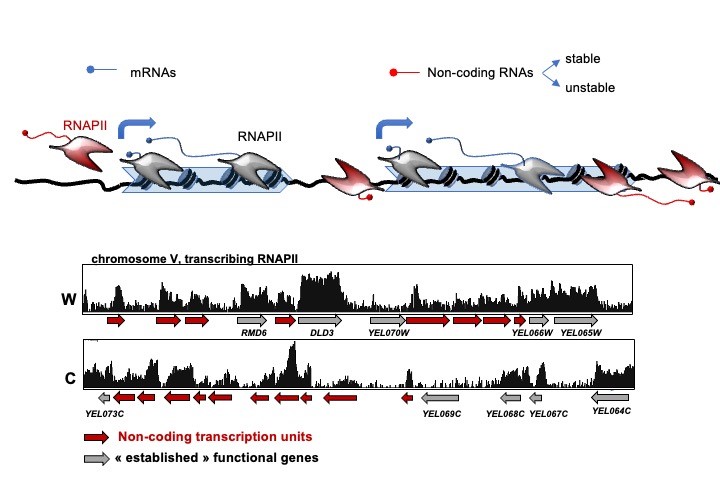
The extended distribution of non-coding transcription events needs to be controlled to avoid potentially disruptive interference with the expression of neighboring genes and other DNA-associated processes. Transcription termination has important roles in this context (see our review: Porrua & Libri, 2015), and studying the mechanisms involved in this process is one of our main interests. Transcription termination at the 3' end of protein-coding genes depends on a conserved multi-subunit complex called the CPF-CF complex. In budding yeast, the NNS complex, composed of the RNA-binding proteins Nrd1 and Nab3, and the helicase Sen1 terminates pervasive transcription events but also transcription of functional non-coding RNA genes such as snoRNAs. Moreover, the NNS complex promotes the degradation of non-coding RNAs by the nuclear exosome and its cofactor the TRAMP complex (figure 2).
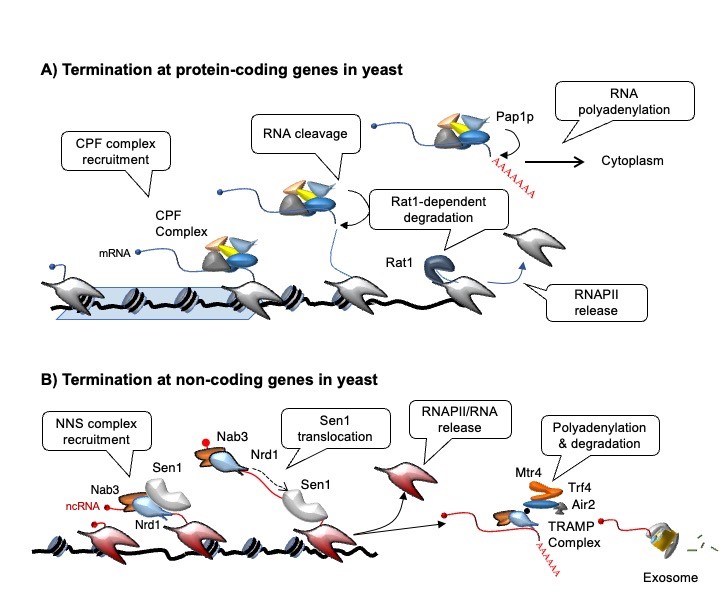
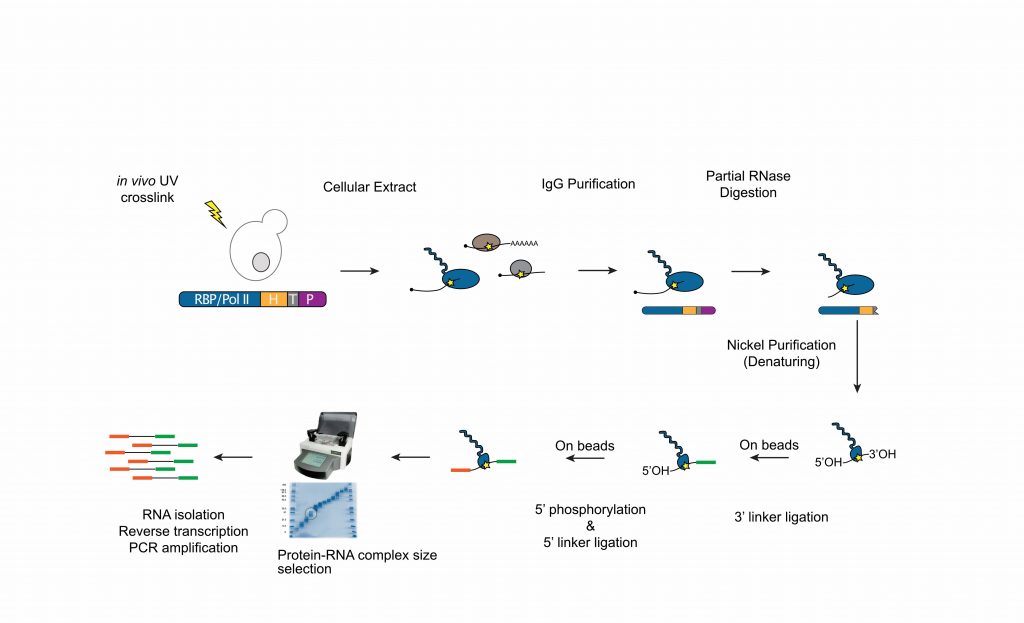
During the past years, we have extensively characterized the function of the NNS-complex. However, many aspects of the mechanisms of termination remain unclear, both for non-coding and mRNA coding genes. We use a technique that allows detecting in vivo the position of the RNAPII with single-nucleotide resolution (CRAC, Crosslinking Analysis of cDNAs, figure 3, Bohnsack et al., 2012; Challal et al., 2022) to generate transcription maps in mutants of the NNS and the CPF-CF pathways.
.
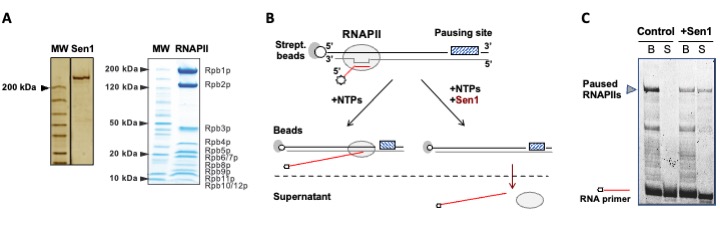
This allows comparative analyses of the different termination pathways and provides some information on the mechanism (together with some unexpected findings!). We like to combine these high-resolution genome-wide analyses with in vitro biochemical (figure 4) and structural approaches to investigate the mechanisms of transcription termination and the interplay between the different termination pathways (as a typical example see our recent publication: Xie et al, 2022).
AXIS 2:
Analysis of the impact of non-coding transcription in gene expression in different physiological conditions
(PI : D. Libri & O. Porrua)
Non-coding transcription events can regulate gene expression by affecting the function of promoters of neighboring genes in the sense or antisense orientation. The regulatory potential of non-coding transcription has been generally neglected, mostly because many previous analyses relied on detection of the RNA as proxy for transcription and ncRNAs produced by potentially regulatory transcription events are often unstable and not easily detected in wild type cells. High resolution and directional mapping the transcribing RNAPII allows circumventing this problem.
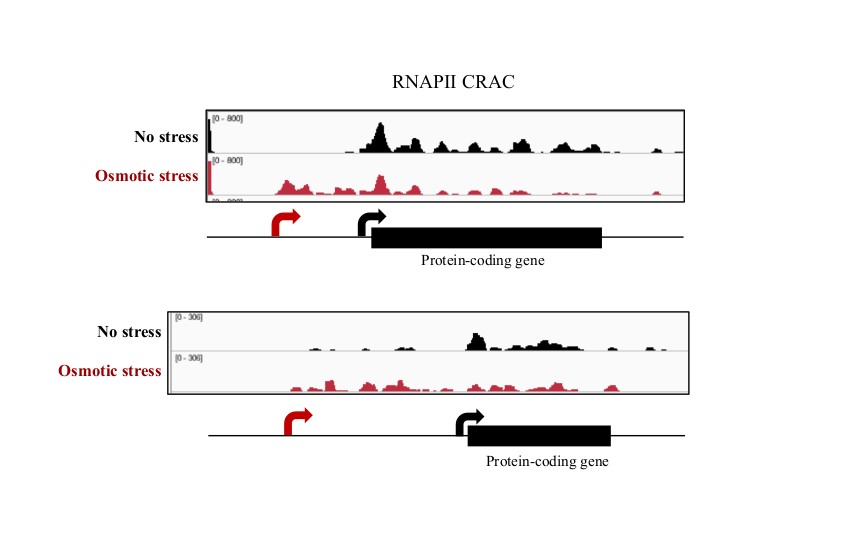
We are interested in further exploring pathways of regulation by non-coding transcription in different physiological and stress conditions. We detected many novel ncRNA transcription events in these conditions, some of which derive from activated bidirectional promoters, some from “solo” non-coding transcription units and some from decreased transcription termination efficiency (see our recent publication Haidara et al, 2022; and figure 5). We are pursuing the study of the impact of non-coding transcription in gene expression using a variety of approaches and bioinformatic tools.
AXIS 3 :
Characterization of the mechanisms responsible for the resolution of transcription-replication conflicts
(PI : D. Libri)
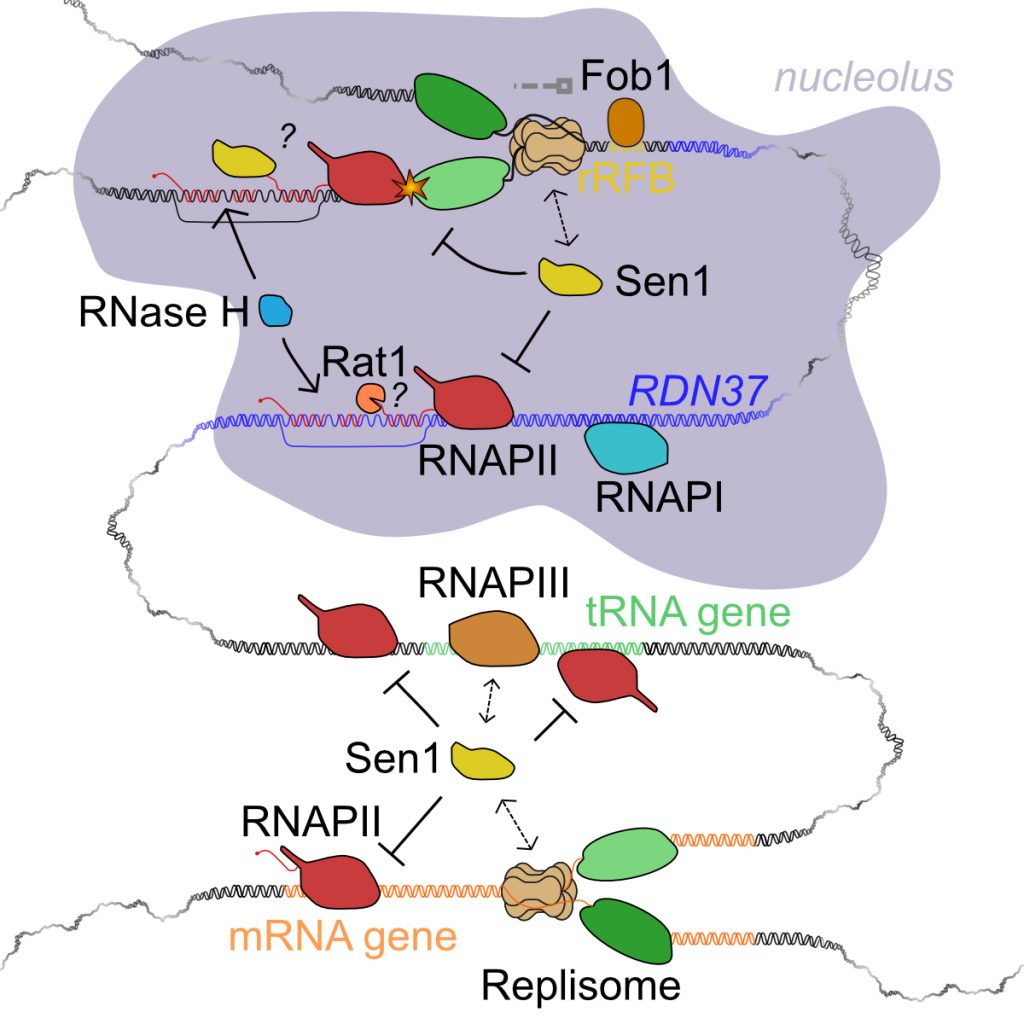
The existence of transcription events that transcend the limits of annotated canonical genes is a major challenge for the cohabitation of transcription and other DNA-associated events such as replication. We have been interested in the recent past the impact that pervasive transcription has on the function of yeast replication origins (see Candelli et al, 2018). We are now directing our interests to the conflicts that are generated by RNAPII transcription. In a series of recent studies (Aiello et al., 2022; Appanah et al., 2020) we have analyzed the relationships of RNAPII transcription with replication and transcription events mediated by other polymerases. We have demonstrated, in collaboration with the de Piccoli, Pasero and Palancade labs, that Sen1, besides and independently of its role in terminating non-coding transcription, has a capital role in controlling transcription-replication and transcription-transcription conflicts. We have shown that Sen1 removes RNAPIIs that collide with the replisome or with transcription of other polymerases, thus qualifying as a master regulator of genomic crowding (Figure 6).
When conflicts occur, the nascent RNA might hybridize to the template DNA strand, forming structures called R-loops. These structures can generate DNA damage and be genotoxic, and are generally removed by RNases H that degrade the RNA portion of the hybrid. Sen1 is also believed to play a role in limiting the formation of R-loops or resolving them once they form. We studied the role of R-loops and RNases H in regions of conflicts, and described a novel method for high resolution R-loop detection. We will pursue the studies of the mechanism of conflict resolution, the roles of RNases H and R-loops in these processes and the impact in genome stability.
AXIS 4 :
Study of the molecular function of human senataxin and its involvement in neurodegeneration
(PI : O.Porrua)
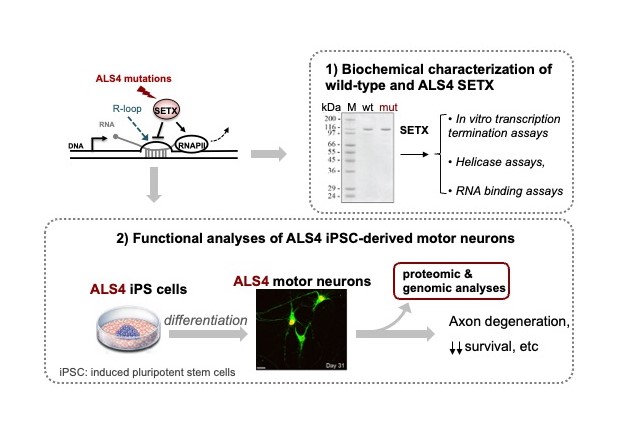
The human homologue of Sen1, senataxin (SETX), has attracted much attention because of its connection with two neurodegenerative diseases. Recessive loss-of-function SETX mutations have been associated with Ataxia with Oculomotor Apraxia type 2 (AOA2), whereas dominant gain-of-function mutations in SETX are linked to a juvenile form of Amyotrophic Lateral Sclerosis (ALS) dubbed ALS4. As Sen1, SETX has been assigned a role in transcription termination as well as in R-loop resolution, however the precise role of SETX in these processes has remained poorly understood because of the absence of biochemical data on the properties and activities of SETX. In addition, a systematic identification of the protein interactors and the targets of SETX is missing. To fill these gaps, we have benefitted from our expertise and the tools we have developed for the functional characterization of the yeast homologue of SETX to elucidate the molecular function of SETX. In collaboration with M. Sebesta and R. Stefl we have recently purified the catalytic domain of SETX and, using a variety of in vitro biochemical assays we have shown for the first time that SETX is a bona fide R-loop resolving helicase and transcription termination factors (see our recent preprint Hasanova et al, 2022, https://www.biorxiv.org/content/10.1101/2022.08.25.505353v1).
In addition, we are currently addressing the molecular basis of SETX-associated ALS in collaboration with S. Nedelec (IFM, Paris). To this end, we have generated human motor neurons from induced pluripotent stem cells harbouring ALS4 mutations and we are employing a variety of approaches to investigate the impact of these mutations on motor neuron physiology. Next, we will combine biochemical, proteomic and genomic approaches to unveil the deregulations responsible for motor neuron impairment in ALS4 (figure 7).
Recent Publications :
1- Aiello, U., Challal D., Wentzinger, G., Lengronne, A., Appanah, R., Pasero, P., Palancade, B. and Libri, D*. (2022). Sen1 is a master regulator of transcription-driven conflicts. Cell S1097-2765(22)00604-9. PMID: 35839782
This work was highlighted by the CNRS
2- Xie, J., Aiello, U., Clement, Y., Haidara, N., Girbig, M., Schmitzova, J., Pena, P. Müller, C.W., Libri, * and Porrua, O*. An integrated model for termination of RNA polymerase III transcription. Sci Adv. 8(28):eabm9875. PMID: 35857496
This work was highlighted by the CNRS
3- Han, Z., Jasnovidova, O., Haidara, N., Tudek, A., Kubicek, K., Libri, D., Stefl, R. and Porrua, O* (2020). Termination of non-coding transcription in yeast relies on both an RNA Pol II CTD-interaction domain and a CTD-mimicking region in Sen1. EMBO J 39(7):e101548. PMID: 32107786
This work was recommended in Faculty Opinions.
4- Challal, D., Barucco, M., Kubik, S., Feuerbach, F., Candelli, T., Geoffroy, H., Benaksas, C., Shore, D., and Libri, D*. (2018). General Regulatory Factors Control the Fidelity of Transcription by Restricting Non-coding and Ectopic Initiation. Cell 72, 955-969.e7. PMID: 30576657
List of all the publication :
*co-corresponding authors
Reviews in italic
Hasanova, Z., Klapstova, V., Porrua*, O., Stefl*, R., and Sebesta*, M. (2022). Human senataxin is a bona fide R-loop resolving enzyme and transcription termination factor. 2022.08.25.505353. https://doi.org/10.1101/2022.08.25.505353.
Girbig, M., Xie, J., Grötsch, H., Libri, D., Porrua, O., and Müller, C.W. (2022). Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals. Cell Reports 40. https://doi.org/10.1016/j.celrep.2022.111316.
Villa, T. and Porrua, O (2022). Pervasive transcription: a controlled risk. FEBS J. doi: 10.1111/febs.16530. PMID: 35587776
Haidara, N., Giannini, M. and Porrua, O (2022). Modulated termination of non-coding transcription partakes in the regulation of gene expression. Nucleic Acids Res 50(3):1430-1448. PMID: 35037029.
Challal, D., Colin, J., Villa, T., and Libri, D. (2022). A Modified Cross-Linking Analysis of cDNAs (CRAC ) Protocol for Detecting RNA-Protein Interactions and Transcription at Single-Nucleotide Resolution. Methods Mol Biol 2477, 35–55. https://doi.org/10.1007/978-1-0716-2257-5_3.
Scutenaire, J., Plassard, D., Matelot, M., Villa, T., Zumsteg, J., Libri, D., and Séraphin, B. (2022). The S. cerevisiae m6A-reader Pho92 promotes timely meiotic recombination by controlling key methylated transcripts. Nucleic Acids Res gkac640. https://doi.org/10.1093/nar/gkac640.
Aiello, U., Challal, D., Wentzinger, G., Lengronne, A., Appanah, R., Pasero, P., Palancade, B., and Libri, D. (2022). Sen1 is a key regulator of transcription-driven conflicts. Mol Cell 82, 2952-2966.e6. https://doi.org/10.1016/j.molcel.2022.06.021.
Xie, J., Aiello, U., Clement, Y., Haidara, N., Girbig, M., Schmitzova, J., Pena, V., Müller, C.W., Libri, D., and Porrua, O. (2022). An integrated model for termination of RNA polymerase III transcription. Sci Adv 8, eabm9875. https://doi.org/10.1126/sciadv.abm9875.
Villa, T., Barucco, M., Martin-Niclos, M.-J., Jacquier, A., and Libri, D. (2020). Degradation of Non-coding RNAs Promotes Recycling of Termination Factors at Sites of Transcription. Cell Rep 32, 107942.
Porrua, O. (2020). Purification and in vitro analysis of the exosome cofactors Nrd1-Nab3 and Trf4-Air2. Methods Mol Biol 2062:277-289. PMID: 31768982
Begley, V., Jordán-Pla, A., Peñate, X., Garrido-Godino, A.I., Challal, D., Cuevas-Bermúdez, A., Mitjavila, A., Baruc co, M., Gutiérrez, G., Singh, A., Alepuz, P., Navarro, F., Libri* , D., Perez-Ortin*, JE., Chavez*, D. (2020). Xrn1 influence on gene transcription results from the combination of general effects on elongating RNA pol II and gene-specific chromatin configuration. RNA Biol 1–14.
Appanah, R., Lones, E.C., Aiello, U., Libri, D., and De Piccoli, G. (2020). Sen1 Is Recruited to Replication Forks via Ctf4 and Mrc1 and Promotes Genome Stability. Cell Rep 30, 2094-2105.e9.
Han, Z., Jasnovidova, O., Haidara, N., Tudek, A., Kubicek, K., Libri, D., Stefl, R., and Porrua, O. (2020). Termination of non-coding transcription in yeast relies on both an RNA Pol II CTD interaction domain and a CTD-mimicking region in Sen1. EMBO J. e101548.
Kubik, S., Bruzzone, M.J., Challal, D., Dreos, R., Mattarocci, S., Bucher, P., Libri, D., and Shore, D. (2019). Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat. Struct. Mol. Biol. 26, 744–754.
Wang, S., Han, Z., Libri, D., Porrua, O., and Strick, T.R. (2019). Single-molecule characterization of extrinsic transcription termination by Sen1 helicase. Nat Commun 10, 1545.
Han, Z. and Porrua, O. (2018). Helicases as transcription termination factors: Different solutions for a common problem. Transcription. 9(3):152-158. PMID: 28886303
Candelli, T., Gros*, J., and Libri*, D. (2018). Pervasive transcription fine-tunes replication origin activity. Elife 7.
Challal, D., Barucco, M., Kubik, S., Feuerbach, F., Candelli, T., Geoffroy, H., Benaksas, C., Shore, D., and Libri, D. (2018). General Regulatory Factors Control the Fidelity of Transcription by Restricting Non-coding and Ectopic Initiation. Mol. Cell 72, 955-969.e7.
Candelli, T., Challal, D., Briand, J.-B., Boulay, J., Porrua, O., Colin, J., and Libri, D. (2018). High-resolution transcription maps reveal the widespread impact of roadblock termination in yeast. EMBO J. 37.
Luciano, P.°, El-kaoutari°, A., Challal° D., Jeon J., Candelli, T., Jourquin, F., Kim, J.*, Libri, D.*, and Geli, V.* (2017) Binding to RNA in vivo regulates Set1 function, Cell Discov 3, 17040.
Simonetti, F., Candelli,T., Leon, S., Libri, D., Rougemaille, M. (2017) Ubiquitination-dependent control of sexual differentiation in fission yeast. Elife 6
Leonaitė, B., Han, Z., Basquin, J., Bonneau, F., Libri, D., Porrua*, O., and Conti*, E. (2017). Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family. EMBO J.
Han, Z., Libri, D., and Porrua, O. (2017). Biochemical characterization of the helicase Sen1 provides new insights into the mechanisms of non-coding transcription termination. Nucleic Acids Res. 45, 1355–1370.
Babour, A., Shen, Q., Dos-Santos, J., Murray, S., Gay, A., Challal, D., Fasken, M., Palancade, B., Corbett, A., Libri, D., Mellor, J. and Dargemont, C. (2016). The Chromatin Remodeler ISW1 Is a Quality Control Factor that Surveys Nuclear mRNP Biogenesis. Cell 167, 1201–1214.e15.
Porrua, O., Boudvillain, M., and Libri, D. (2016). Transcription Termination: Variations on Common Themes. Trends Genet. 32, 508–522.
Libri, D. (2015). Sleeping Beauty and the Beast (of pervasive transcription). RNA 21, 678–679.
Porrua, O., and Libri, D. (2015a). Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 16, 190–202.
Porrua, O., and Libri, D. (2015b). Characterization of the mechanisms of transcription termination by the helicase Sen1. Methods Mol. Biol. 1259, 313–331.
Tudek, A., Candelli, T., and Libri, D. (2015). Non-coding transcription by RNA polymerase II in yeast: Hasard or nécessité? Biochimie. 117, 28-36
Zhang, W., Collinet, B., Graille, M., Daugeron, M.-C., Lazar, N., Libri, D., Durand, D., and van Tilbeurgh, H. (2015). Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res. 43, 3358–3372.
Libri, D. (2015). Endless Quarrels at the End of Genes. Mol. Cell 60, 192–194.
Colin, J., Candelli, T., Porrua, O., Boulay, J., Zhu, C., Lacroute, F., Steinmetz, L.M., and Libri, D. (2014). Roadblock termination by reb1p restricts cryptic and readthrough transcription. Mol. Cell 56, 667–680.
Tudek*, A., Porrua*, O., Kabzinski, T., Lidschreiber, M., Kubicek, K., Fortova, A., Lacroute, F., Vanacova, S., Cramer, P., Stefl, R. and Libri, D. (2014). Molecular basis for the dual role of Nrd1p CID in transcription termination and non-coding RNA degradation. Mol. Cell, 55, 467-81
Mouaikel, J., Causse, S.Z., Rougemaille, M., Daubenton-Carafa, Y., Blugeon, C., Lemoine, S., Devaux, F., Darzacq, X., and Libri, D. (2013). High-Frequency Promoter Firing Links THO Complex Function to Heavy Chromatin Formation. Cell Rep 5, 1082-1094.
Lenstra*, T.L., Tudek*, A., Clauder, S., Xu, Z., Pachis, S.T., van Leenen, D., Kemmeren, P., Steinmetz, L.M., Libri*, D., and Holstege*, F.C. (2013). The Role of Ctk1 Kinase in Termination of Small Non-Coding RNAs. PLoS One 8, e80495.
Jensen, T.H., Jacquier, A., and Libri, D. (2013). Dealing with pervasive transcription. Mol Cell 52, 473-484.
Porrua, O., and Libri, D. (2013). A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol 20, 884-891.
Porrua, O., and Libri, D. (2013). RNA quality control in the nucleus: the Angels’ share of RNA. Biochim. Biophys. Acta 1829, 604–611.
Rougemaille, M., Braun, S., Coyle, S., Dumesic, P.A., Garcia, J.F., Isaac, R.S., Libri, D., Narlikar, G.J., and Madhani, H.D. (2012). Ers1 links HP1 to RNAi. Proc. Natl. Acad. Sci. U.S.A. 109, 11258–11263.
Porrua, O., Hobor, F., Boulay, J., Kubicek, K., D'Aubenton-Carafa, Y., Gudipati, R.K., Stefl, R., and Libri, D. (2012). In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. Embo J 31, 3935-3948.
Gudipati, R.K., Xu, Z., Lebreton, A., Seraphin, B., Steinmetz, L.M., Jacquier, A., and Libri, D. (2012). Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48, 409-421.
Rougemaille, M., and Libri, D. (2011). Control of cryptic transcription in eukaryotes. Adv Exp Med Biol 702, 122-131.
Daugeron, M.C., Lenstra, T.L., Frizzarin, M., El Yacoubi, B., Liu, X., Baudin-Baillieu, A., Lijnzaad, P., Decourty, L., Saveanu, C., Jacquier, A., Holstege, F.C.P., de Crécy-Lagard, V., van Tilbeurgh, H. and Libri, D. (2011). Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res 39, 6148-6160.
Colin, J., Libri, D., and Porrua, O. (2011). Cryptic transcription and early termination in the control of gene expression. Genet Res Int 2011, 653494.
Assenholt, J., Mouaikel, J., Saguez, C., Rougemaille, M., Libri, D., and Jensen, T.H. (2011). Implication of Ccr4-Not complex function in mRNA quality control in Saccharomyces cerevisiae. Rna 17, 1788-1794.
Libri, D. (2010). Nuclear poly(a)-binding proteins and nuclear degradation: take the mRNA and run? Mol Cell 37, 3-5.
Honorine, R., Mosrin-Huaman, C., Hervouet-Coste, N., Libri, D., and Rahmouni, A.R. (2010). Nuclear mRNA quality control in yeast is mediated by Nrd1 co-transcriptional recruitment, as revealed by the targeting of Rho-induced aberrant transcripts. Nucleic Acids Res 39, 2809-2820.
Colin, J., Libri, D., and Villa, T. (2010). Sex matters in the birth of genes. Cell Res 20, 499-501.
Villa, T., Rougemaille, M., and Libri, D. (2008). Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: Tilting the balance to shape the transcriptome. Biochim Biophys Acta 1779, 524-531.
Thiebaut, M., Colin, J., Neil, H., Jacquier, A., Seraphin, B., Lacroute, F., and Libri, D. (2008). Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31, 671-682.
Schmid, M., Rougemaille, M., Libri, D., and Jensen, T.H. (2008). Chapter 10. Estimating nuclear mRNA decay in Saccharomyces cerevisiae. Methods Enzymol 449, 205-219.
Rougemaille, M., Villa, T., Gudipati, R.K., and Libri, D. (2008). mRNA journey to the cytoplasm: attire required. Biol Cell 100, 327-342.
Rougemaille, M., Dieppois, G., Kisseleva-Romanova, E., Gudipati, R.K., Lemoine, S., Blugeon, C., Boulay, J., Jensen, T.H., Stutz, F., Devaux, F., and Libri, D. (2008). THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association. Cell 135, 308-321.
Libri, D. (2008). Quality control: Cell and the city. Biochim Biophys Acta 1779, 523.
Hecker, A., Lopreiato, R., Graille, M., Collinet, B., Forterre, P., Libri, D., and van Tilbeurgh, H. (2008). Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. Embo J 27, 2340-2351.
Gudipati, R.K., Villa, T., Boulay, J., and Libri, D. (2008). Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol 15, 786-794.
Assenholt, J., Mouaikel, J., Andersen, K.R., Brodersen, D.E., Libri*, D., and Jensen*, T.H. (2008). Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. Rna 14, 2305-2313.
Rougemaille, M., Gudipati, R.K., Olesen, J.R., Thomsen, R., Seraphin, B., Libri*, D., and Jensen*, T.H. (2007). Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. Embo J 26, 2317-2326.
Chauveau, F., Aissouni, Y., Hamm, J., Boutin, H., Libri, D., Duconge, F., and Tavitian, B. (2007). Binding of an aptamer to the N-terminal fragment of VCAM-1. Bioorg Med Chem Lett 17, 6119-6122.
Thiebaut, M., Kisseleva-Romanova, E., Rougemaille, M., Boulay, J., and Libri, D. (2006). Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell 23, 853-864.
Pestourie, C., Cerchia, L., Gombert, K., Aissouni, Y., Boulay, J., De Franciscis, V., Libri, D., Tavitian, B., and Duconge, F. (2006). Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides 16, 323-335.
Kisseleva-Romanova, E., Lopreiato, R., Baudin-Baillieu, A., Rousselle, J.C., Ilan, L., Hofmann, K., Namane, A., Mann, C., and Libri, D. (2006). Yeast homolog of a cancer-testis antigen defines a new transcription complex. Embo J 25, 3576-3585.
Cerchia, L., D'Alessio, A., Amabile, G., Duconge, F., Pestourie, C., Tavitian, B., Libri, D., and de Franciscis, V. (2006). An autocrine loop involving ret and glial cell-derived neurotrophic factor mediates retinoic acid-induced neuroblastoma cell differentiation. Mol Cancer Res 4, 481-488.
Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin*, B., Libri*, D. and Jacquier* A. (2005). Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725-737. * co-corresponding authors.
Olesen, J.R., Libri, D., and Jensen, T.H. (2005). A link between transcription and mRNP quality in Saccharomyces cerevisiae. RNA Biol 2, 45-48.
Jensen, T.H., Boulay, J., Olesen, J.R., Colin, J., Weyler, M., and Libri, D. (2004). Modulation of transcription affects mRNP quality. Mol Cell 16, 235-244.
Thomsen, R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. (2003). Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. Rna 9, 1049-1057.
Jensen, T.H., Dower, K., Libri, D., and Rosbash, M. (2003). Early formation of mRNP: license for export or quality control? Mol Cell 11, 1129-1138.
Cerchia, L., Libri, D., Carlomagno, M.S., and de Franciscis, V. (2003). The soluble ectodomain of RetC634Y inhibits both the wild-type and the constitutively active Ret. Biochem J 372, 897-903.
Libri, D., Duconge, F., Levy, L., and Vinauger, M. (2002). A role for the Psi-U mismatch in the recognition of the 5' splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J Biol Chem 277, 18173-18181.
Libri*, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen*, T.H. (2002). Interactions between mRNA export commitment, 3'-end quality control, and nuclear degradation. Mol Cell Biol 22, 8254-8266.
Cerchia, L., Hamm, J., Libri, D., Tavitian, B., and de Franciscis, V. (2002). Nucleic acid aptamers in cancer medicine. FEBS Lett 528, 12-16.
Libri, D., Graziani, N., Saguez, C., and Boulay, J. (2001). Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15, 36-41.
Jensen, T.H., Boulay, J., Rosbash, M., and Libri, D. (2001). The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol 11, 1711-1715.



