Notre équipe s’intéresse au contrôle de l’expression des gènes dans les cellules mammifères avec un intérêt particulier pour la différenciation des cellules T. Un aspect important de notre recherche est l’étude du rôle des résidus du Domaine Carboxy-Terminal (CTD) de l’ARN Polymerase II (Pol II) dans la régulation transcriptionnelle (axe 1).
Au cours de la différenciation, d’importants changements de l’expression des gènes associés à des modifications épigénétiques du génome se produisent dans les cellules souches, leur permettant d’évoluer vers leur destin cellulaire. Ces changements sont couplés à des transitions dans les fonctions et les propriétés des cellules T. Les promoteurs et les enhancers des unités géniques codantes ou non-codantes sont des modules génomiques essentiels à la mise en place de ces transitions (axes 2 et 3). Le décryptage de leur mécanisme d’action est l’un des défis que nous souhaitons relever et lie entre eux nos trois axes de recherche.
Axe 1 Rôle des résidus du domaine CTD de l’ARN Polymerase II dans le cycle transcriptionnel.
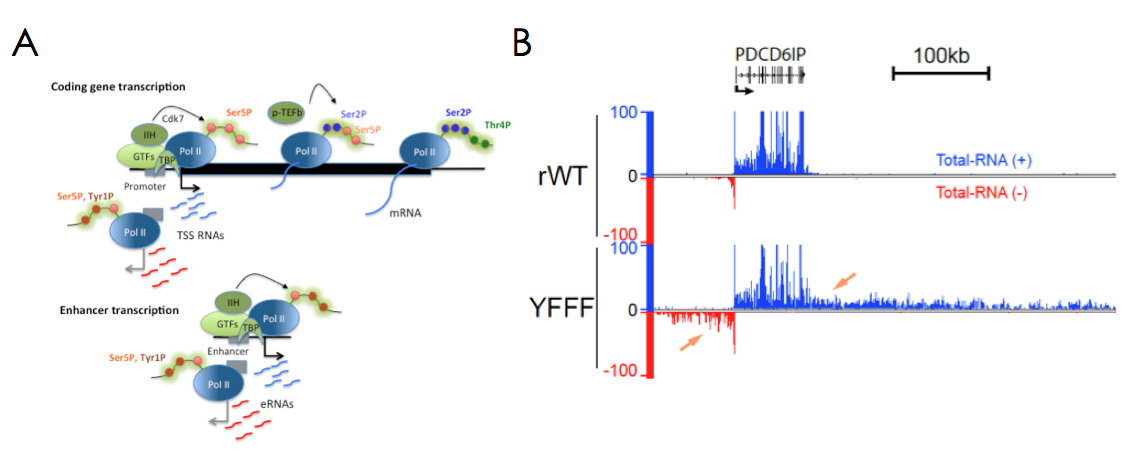
Le domaine CTD de Pol II et ses modifications post-traductionnelles jouent un rôle crucial dans les étapes de recrutement, d’initiation et d’élongation de la transcription. Ils sont également importants pour l’épissage, le traitement et la stabilité des ARNs. Le CTD est composé d’une répétition du motif peptidique Y1S2P3T4S5P6S7 dont les résidus Sérine, Thréonine et Tyrosine peuvent être modifiés par phosphorylation. Les résidus Ser5P et Ser2P sont bien décrits pour leur rôle dans l’initiation et l’élongation alors que la modification Ser7P est vraisemblablement impliquée dans la maturation des snRNAs. Nous avons montré que les résidus Tyr1P et Thr4P sont probablement liés à la transcription bidirectionnelle aux promoteurs et à la terminaison transcriptionnelle, respectivement (Figure1A ;Descostes, Elife 2014 May 9:e02105, Hintermair, EMBO J. 2012 31, 2784). Nos travaux plus récents sur la Tyrosine1 du CTD ont mis en évidence son implication critique dans le contrôle de la terminaison transcriptionnelle en amont des promoteurs (transcription divergente) et après la fin des régions codantes (Shah, Maqbool et al, Mol. Cell 2018 69, 48). Un mutant du CTD (YFFF) où la Tyr1 est remplacé par un résidu Phe présente un phénotype ‘pervasif’ exceptionnel, où la Pol II peut transcrire de manière aberrante plusieurs centaines de kb en amont ou en aval des gènes (Figure1B). Ce mutant perd également l’interaction avec les complexes Médiateur et Intégrateur, suggérant leur association avec le processus de terminaison.
Figure 1: Modifications du CTD et transcription pervasive d’un mutant de la Tyr1.
A- Modèle du cycle transcriptionnel intégrant les modifications du CTD. Nos travaux ont permis de mettre en évidence les localisations des résidus Tyr1P aux promoteurs et aux enhancers et de la Thr4P à la fin et après les extrémités 3’ des gènes (Descostes, Elife 2014 May 9:e02105, Hintermair, EMBO J. 2012 31, 2784).
B- Les résidus Tyr1 du CTD sont impliqués dans la terminaison de la transcription chez les mammifères. Exemple d’un phénotype de transcription aberrante d’une Polymérase mutée pour la tyrosine (YFFF) avant et après les extrémités du gène PDCD6IP (Shah, Maqbool et al, Mol. Cell 2018 69, 48).
Axe 2Rôle des enhancers au cours de la différenciation des cellules T.
Nous avons montré dans le passé que les enhancers transcrits sont des marqueurs d’identité et d’expression tissu-spécifique dans les cellules T en développement (Koch, NSMB 2011 18, 956). Nous avons également démontré l’existence de Plateformes d’Initiation de la Transcription (TIPs), liées aux promoteurs et aux enhancers, dont les caractéristiques sont très proches des ‘Super-enhancers’ décrits plus récemment. Nous étudions aussi des facteurs de transcription spécifiques et avons mis en évidence le rôle critique du facteur Ets1 dans l’engagement vers le lignage T (Cauchy, NAR 2016 44, 3567). Ets1 peut également lier les régions ouvertes et fermées de la chromatine, comme le ferait un facteur pionnier. Dans d’autres travaux collaboratifs nous avons contribué à l’établissement d’une nouvelle procédure de mesure d’activité enhancer à l’échelle du génome (Vanhille, Nat. Com. 2015 6, 6905) et à montrer que les enhancers spécifiques au lignage activent des réseaux de gènes liés à l’auto-renouvellement dans les macrophages et les cellules ES (Soucie, Science 2016 12, 351).
Nos travaux actuels se concentrent sur l’analyse de la dynamique des enhancers au cours de la différenciation des cellules T CD4, en adressant en particulier la question- pourquoi les unités géniques utilisent-elles différents éléments régulateurs au cours du développement et de la différenciation ?-
Nos études sur les éléments régulateurs s’appliquent également au cancer, en particulier aux modèles de leucémies lymphoïdes aigues T (Navarro, Nat. com. 2015 6:6905). Nous avons ainsi montré qu’une petite insertion en amont de l’oncogène Tal1 conduit à son activation anormale dans les cellules T. Cette insertion conduit à la neo-génése d’un enhancer actif et à la dé-répression du gène Tal1 par le complexe Polycomb. Nous poursuivons l’étude de ce mécanisme sur d’autres oncogènes.
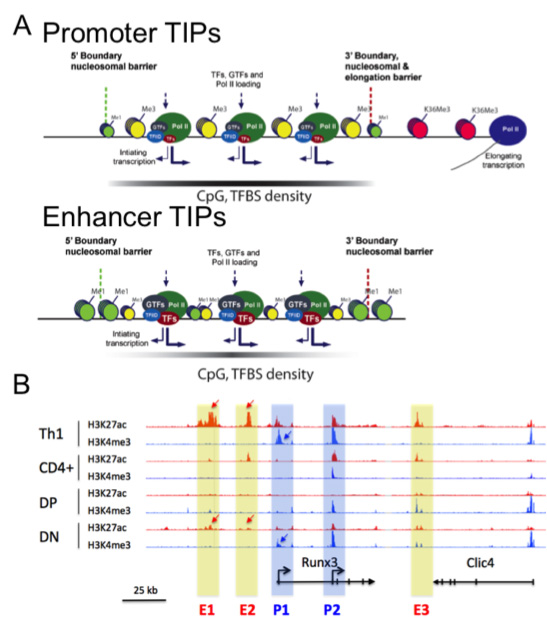
Figure 2: Dynamique des enhancers et des promoteurs dans les cellules T en développement.
A- Les plateformes d’Initiation de la Transcription (TIPs) sont de larges zones, présentes aux promoteurs et aux enhancers actifs et fortement associées à l’identité cellulaire. Les TIPs aux promoteurs et aux enhancers ont une signature épigénétique proche mais les enhancers ne présentent pas de marque d’élongation de la transcription (Koch, NSMB 2011 18, 956).
B- Dynamique des marques épigénétiques actives aux promoteurs (H3K4me3) et aux enhancers (H3K27ac) au cours de la différenciation des cellules T murines au locus Runx3. Les flèches indiquent les régions régulatrices présentant une forte dynamique aux enhancers putatifs (rectangles jaunes) ou aux promoteurs (rectangles bleus) de Runx3 (données non publiées).
Axe 3Déterminant des structures promoteurs dans les cellules mammifères.
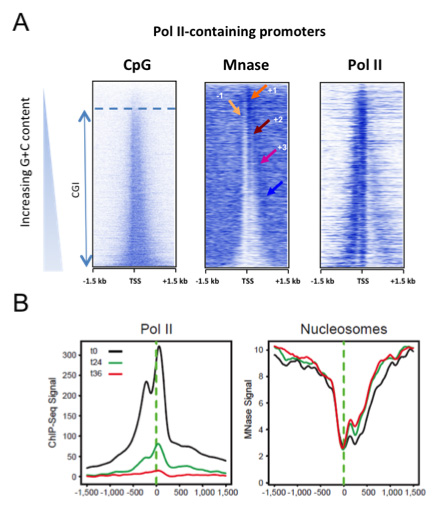
Depuis plusieurs décennies, la question des éléments de séquence critiques pour la mise en place d’un promoteur reste ouverte. L’élément dit ‘TATA-box’ est peu représenté dans la plupart des promoteurs. A l’inverse, les ilots CpG (CGIs), riches en nucléotides G/C sont présents dans environ ¾ des promoteurs mammifères. Nous avons montré que les CGIs sont essentiels à l’ouverture de la chromatine et à la déplétion nucléosomale des promoteurs. Cette propriété est indépendante du niveau de transcription des gènes (Fenouil, Gen. Res. 2012 22, 2399). Nous poursuivons ces travaux afin de mieux comprendre et disséquer les mécanismes moléculaires à l’origine de cette ouverture. Dans d’autres études, nous avons contribué à analyser la déplétion nucléosomale liée aux CGIs dans la réplication (Cayrou, Gen. Res. 2015, 25:1873) et dans l’expression de long transcrits non-codants situés immédiatement en amont des gènes (Lepoivre, BMC Gen. 2013 23;14:914). Nous proposons que des structures secondaires de l’ADN sont à l’œuvre dans les CGIs aux promoteurs, expliquant l’ouverture de la chromatine en présence ou en absence de transcription. Nous développons des procédures pour isoler et caractériser de telles structures in vivo, via des stratégies couplées au séquençage à haut débit.
Figure 3: Les promoteurs à ilots CpG (CGIs) excluent intrinsèquement les nucléosomes, indépendamment de la transcription (Fenouil, Gen. Res. 2012 22, 2399).
A- Les promoteurs actifs sont classés par niveau de nucleotide CpG croissant (panel gauche, du haut vers le bas). Ce niveau corrèle à la déplétion nucléosomale (panel du milieu).
B- L’inhibition de la transcription ne modifie pas l’ouverture des promoteurs. Des cellules B humaines ont été traitées par de l’a-amanitine aux temps indiqués, résultant en l’inhibition de la transcription (métaprofil ChIP-seq, panel gauche), alors que le niveau de déplétion nucléosomale est peu affecté (MNase-seq, panel droit).