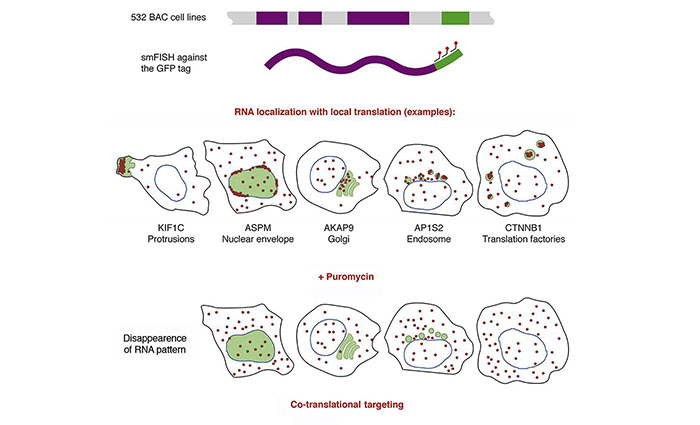
Local translation allows spatial control of gene expression. Here, we performed a dual protein-mRNA localization screen, using smFISH on 523 human cell lines expressing GFP-tagged genes. 32 mRNAs displayed specific cytoplasmic localizations with local translation at unexpected locations, including cytoplasmic protrusions, cell edges, endosomes, Golgi, the nuclear envelope, and centrosomes, the latter being cell-cycle-dependent. Automated classification of mRNA localization patterns revealed a high degree of intercellular heterogeneity. Surprisingly, mRNA localization frequently required ongoing translation, indicating widespread co-translational RNA targeting. Interestingly, while P-body accumulation was frequent (15 mRNAs), four mRNAs accumulated in foci that were distinct structures. These foci lacked the mature protein, but nascent polypeptide imaging showed that they were specialized translation factories. For β-catenin, foci formation was regulated by Wnt, relied on APC-dependent polysome aggregation, and led to nascent protein degradation. Thus, translation factories uniquely regulate nascent protein metabolism and create a fine granular compartmentalization of translation.
Racha Chouaib 1, Adham Safieddine 1, Xavier Pichon 1, Arthur Imbert 1, Oh Sung Kwon, Aubin Samacoits, Abdel-Meneem Traboulsi, Marie-Cécile Robert, Nikolay Tsanov, Emeline Coleno, Ina Poser, Christophe Zimmer, Anthony Hyman, Hervé Le Hir, Kazem Zibara, Marion Peter, Florian Mueller*, Thomas Walter*, Edouard Bertrand*
DOI:https://doi.org/10.1016/j.devcel.2020.07.010
Article highlighted in Dev Cell by Ashley Chin & Eric Lécuyer
Translating Messages in Different Neighborhoods
DOI:https://doi-org.insb.bib.cnrs.fr/10.1016/j.devcel.2020.09.006
Also on the same theme within the Edouard Bertrand Lab IGMM and available on BioRxiv:
Adham Safieddine, Emeline Coleno, Abdel-Meneem Traboulsi, Oh Sung Kwon, Frederic Lionneton, Virginie Georget, Marie-Cécile Robert, Thierry Gostan, Charles Lecellier, Soha Salloum, Racha Chouaib, Xavier Pichon, Hervé Le Hir, Kazem Zibara, Marion Peter, Edouard Bertrand