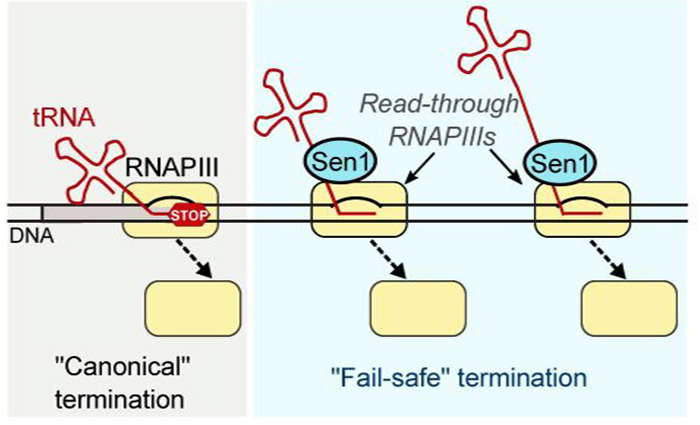
In eukaryotic organisms transcription is carried out by three distinct RNAPs that are specialized in producing different classes of transcripts and seemingly adopt different strategies to terminate transcription. RNA polymerase III (RNAPIII) is responsible for the synthesis of highly abundant transcripts such as transfer RNAs and the 5S ribosomal RNA that are absolutely required for cell proliferation. The traditional model for RNAPIII transcription termination posits that RNAPIII can terminate efficiently and precisely at a specific DNA sequence without the need for any accessory protein. However, this model turned out to be too simplistic. In this study the authors show that the mechanisms driving termination of RNAPIII transcription are more complex and sophisticated than previously proposed.
First, using a state-of-the-art genomic approach allowing the mapping of transcribing RNAPIIIs with single-nucleotide resolution Xie and coauthors observed that a substantial fraction of RNAPIIIs do not respect the “stop” signals at the end of genes and transcribe downstream regions. Next they discovered that termination of RNAPIIIs that have read-though the canonical terminator depends on the interaction of RNAPIII with a highly-conserved protein, the RNA helicase Sen1, which has previously been assigned a major role in transcription termination by RNAPI and RNAPII. Using an in vitro system with highly purified components to assess termination in a controlled manner, they demonstrated that Sen1 can directly induce termination of transcription by RNAPIII. Therefore, this study unveils the existence of a universal mechanism of termination relying on a conserved protein factor that operates on all three eukaryotic RNAPs despite their different properties and subunit composition.
Taken together, these results underscore how organisms have evolved complementary pathways to prevent RNAPs from invading genomic regions beyond their transcribed genes.