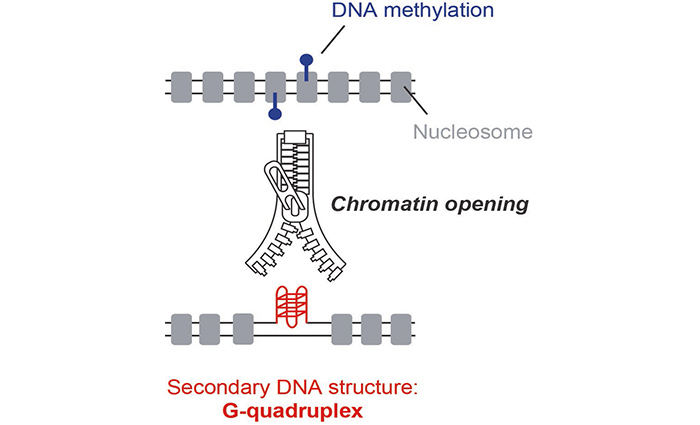
In this work, JC Andrau’s team proposes a new approach for identifying secondary DNA structures, known as G-quadruplexes (G4s), in the nuclear context. This technique, called G4access, uses the properties of a nuclease that digests DNA, under conditions where the G4s resist this digestion and remain protected from the enzyme. After sequencing the digestion residues, it is possible to determine the location of the G4s in the genome (Figure). Data from this work show that G4s are essential determinants for chromatin opening in the genome and are enriched in transcriptionally active regions, although transcription is not strictly required for detection. The G4s observed are also enriched at loci subject to parental imprinting at unmethylated alleles and are antagonist to DNA methylation. Finally, the G4access technique revealed a potential mechanism of action for a G4 ligand at genomic sites.
This study opens up important perspectives for understanding the contribution of DNA secondary structures to genome expression, duplication and maintenance. It also provides a framework for identifying targets for G4 ligands and their potential therapeutic improvements.
These results are published in the journal Nature Genetics.
G4access identifies G-quadruplexes and their associations with open chromatin and imprinting control regions. Esnault C, Magat T, Zine El Aabidine A, Garcia-Oliver E, Cucchiarini A, Bouchouika S, Lleres D, Goerke L, Luo Y, Verga D, Lacroix L, Feil R, Spicuglia S, Mergny JL, Andrau JC. Nat Genet. 2023 Jul 3. doi: 10.1038/s41588-023-01437-4.
https://www.nature.com/articles/s41588-023-01437-4