In cells, genetic information is not expressed continuously, but instead stochastically in an intermittent manner. This phenomenon, called transcriptional noise, plays a key role in many situations in biology, including during embryonic development. In joint articles published with the Bertrand lab (IGMM/IGH) in Nature Communications, the Lagha lab used live cell RNA labeling and innovative mathematical approaches to show that transcriptional noise uses a common mechanism in both HIV-infected cells and Drosophila embryos involving pausing of RNA polymerases near the transcription initiation site.
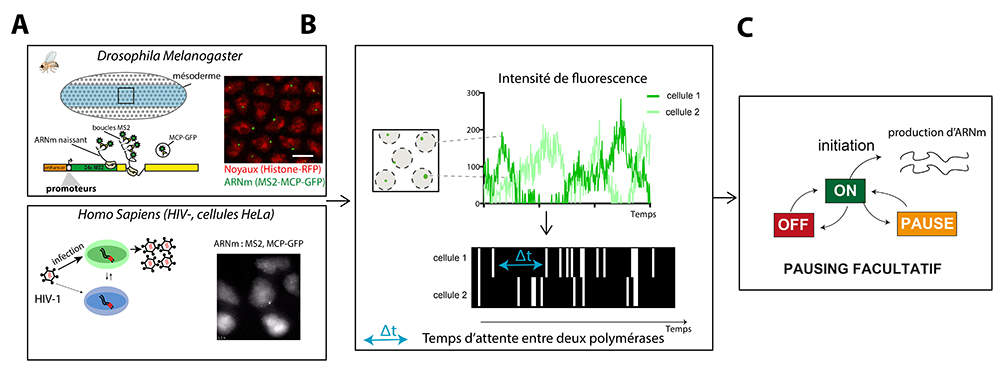
Using Drosophila as a model organism, it is possible to follow mRNA synthesis in real time in a living embryo. In developing embryos, the activation of gene expression takes place discontinuously for different genes, because transcription requires the assembly of many actors whose action is centralized at the level of a particular DNA sequence called a promoter that is unique to each gene. To determine the contribution of promoter sequences to the dynamics of transcription, the Lagha lab combined genetic manipulations with quantitative imaging to reveal the different states of the promoters and their switching kinetics in vivo.
They were able to demonstrate that “TATA box” promoters, identified by a particular conserved DNA sequence, are active very early and operate via a simple stochastic transition between an inactive state and a permissive (active) state, where a polymerase initiates approximately every 10 seconds. For a second type of promoter known as “INR motif” promoters (Initiator), which become active later, an additional inactive state exists which is associated with polymerase pausing near their initiation site. These multiple promoter states switch stochastically at different time scales. In the context of a rapidly developing embryo, this multiscale control contributes to the proper timing and levels of gene expression during the awakening of the zygotic genome.
HIV model: In patients infected with HIV, it is not currently possible to eliminate the virus and people must follow lifelong antiviral treatments. Eradication of HIV is impossible due to latent forms of the virus, in which an infected cell does not express the viral genome and remains undetectable by the immune system. However, these latent viruses can occasionally wake up and restart viral propagation. This produces viral rebounds in patients who stop their antiviral therapy. The mechanisms allowing viral reactivation in latent cells are the subject of numerous studies. One of the mechanisms that limits transcription in latent cells is the arrest of RNA polymerases near the viral promoter. This stop, also called pausing, blocks the initiation of new polymerases and thus inhibits viral expression.Using a method that visualizes viral RNAs in living cells with single-molecule sensitivity, the scientists analyzed viral transcription under conditions mimicking those of latent cells. Surprisingly, these cells display short pulses of viral transcription separated by long periods of inactivity lasting several hours. In contrast to what was previously thought, scientists show that the pause of RNA polymerases near the viral promoter is a stochastic and rare event. Thus, when an arrested polymerase randomly restarts, a whole convoy of new polymerases transcribe the viral genome before a newly-initiating polymerase pauses again. This produces enough viral components to trigger a new infection cycle.
Mathematics decodes cell noise:The scientists analyzed these two biological models using the same mathematical method. This method reconstructs a complete map of polymerase positions over time for each cell. Then, it uses exact solutions of what in mathematics is called an “inverse problem”, to trace the rates of transition between molecular states of the promoter and thus quantify the probability and duration of polymerase pauses. Thus, by analyzing two different biological systems and using a mathematical processing method “making the invisible visible”, the scientists demonstrate that the pause of the RNA polymerases is facultative and stochastic, thus contributing to the intermittency of gene expression.
Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics.
Pimmett V, Dejean M, Fernandez C, Trullo A, Bertrand A, Radulescu O and Lagha M
Nature Communications. 23 juillet 2021. https://doi.org/10.1038/s41467-021-24461-6
Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting
Katjana Tantale, Encar Garcia-Oliver, Marie-Cécile Robert, Adèle L’Hostis, Yueyuxiao Yang, Nikolay Tsanov, Rachel Topno, Thierry Gostan, Alja Kozulic-Pirher, Meenakshi Basu, Vera Slaninova, Jean-Christophe Andrau, Florian Mueller, Eugenia Basyuk, Ovidiu Radulescu, Edouard Bertrand
Nature Communications 23 juillet 2021. https://doi.org/10.1038/s41467-021-24462-5