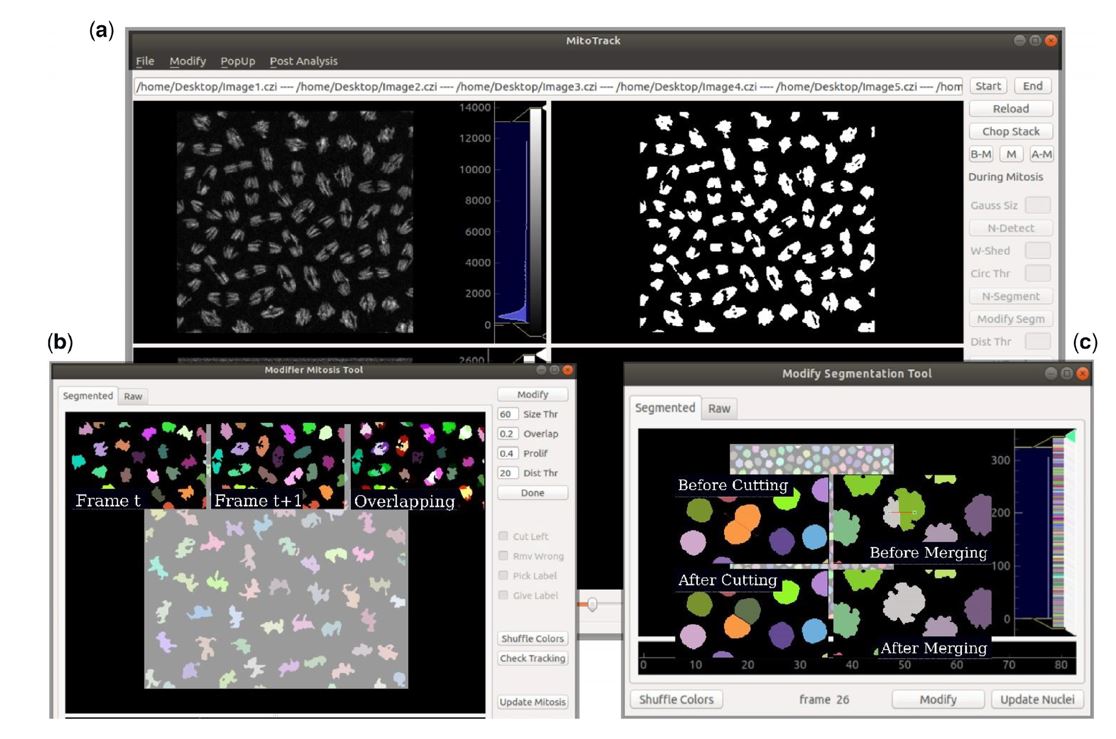
During development, progenitor cells undergo multiple rounds of cellular divisions during which transcriptional programs must be faithfully propagated. To study mitotic inheritance of these programs, it is essential to be able to track transcriptional activities and their transmission from mother to daughter cells in living embryos. Given the stochastic nature of the phenomena, it is necessary to collect and analyze big amount of data to look at the statistical distribution of the results. For this purpose, we developed MitoTrack, a custom build software, developed in PythonTM with a user-friendly graphical user interface (GUI) which provides graphical tools for manual corrections, visualization and check of the results. In order to study the temporal activation of transcriptions in consecutive cell cycles, MitoTrack tracks nuclei across mitosis keeping lineage information and detecting the appearance of transcriptional sites. As a result, we can extract the activation time of each nucleus and its daughters allowing us to study transcriptional memory across mitosis.
Bioinformatics. 2019 Oct 3. pii: btz717. doi: 10.1093/bioinformatics/btz717. [Epub ahead of print]
MitoTrack, a user-friendly semi-automatic software for lineage tracking in living embryos.
Trullo A, Dufourt J, Lagha M.
Unless provided in the caption above, the following copyright applies to the content of this slide: © The Author(s) 2019. Published by Oxford University Press.This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/), which permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited. For commercial re-use, please contact journals.permissions@oup.com