About half of the mammalian genome is constituted of repeated elements among which approximatively 10% are derived from endogenous retroviruses (ERVs). These sequences have often been co-opted by the genome to contribute to endogenous gene expression and specific ERVs are known to be reactivated upon loss of HP1 (Heterochromatin Protein 1), a protein family involved in heterochromatin formation. However, the link between the ERVs, the expression of endogenous genes and the HP1 remains unclear.
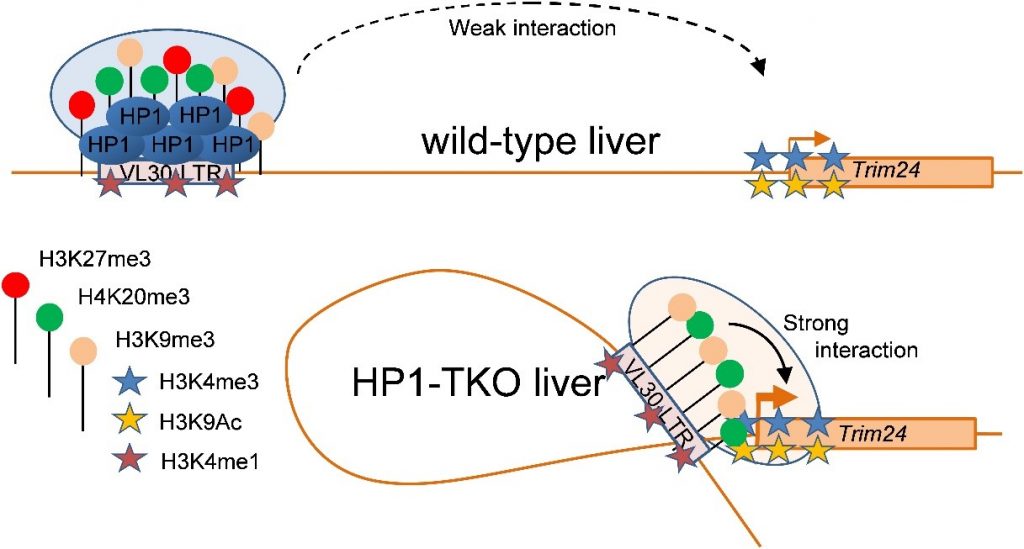
In this work, by studying two ERVs located upstream mouse genes upregulated upon loss of HP1 (Mbd1 and Trim24), the group of Thierry Forné (IGMM) in collaboration with Florence Cammas (IRCM), has shown that some ERVs behave as putative enhancers forming a chromatin loop with promoters of endogenous genes depending on HP1 expression level. These specific ERVs are characterized by HP1-independent (H3K9me3 and H4K20me3) or HP1-dependent (H3K27me3) heterochromatin marks, as well as by the enhancer specific mark H3K4me1, a peculiar combination that might represent a bookmark of HP1-dependent enhancers, derived from ERVs.
Altogether, these results lead to the identification of a new type of transcriptional regulation, involving the HP1-dependent formation of physical loops between specific ERVs and endogenous genes. To our knowledge, this is the first demonstration of a direct long-range interaction between an ERV-derived sequence and the promoter of an endogenous gene in mammals, as well as the first example of a putative enhancer whose activity is regulated by HP1.
Calvet, S. Sallis, N. Saksouk, C. Rebouissou, C. Teyssier, A. Lesne, F. Cammas and T. Forné* – “Endogenous retroviral sequences behave as putative enhancers controlling gene expression through HP1-regulated long-range chromatin interactions” (2022) Cells 11:2392.
https://doi.org/10.3390/cells11152392