The peptidic post-translational modifiers of the Ubiquitin Family (UbL) are reversibly conjugated to thousands of cellular proteins, the activity, function and/or fate of which they modify. In this way, they play a role as important as that of phosphorylation in virtually all cellular processes. In particular, they are essential for the control of gene expression and is disrupted in a variety of disease situations.
In this context, we are studying how UbL, in particular SUMO, notably through their ability to regulate gene expression programs, are involved in the response of Acute Myeloid Leukemia (AML) to therapies, as AML is a hematological malignancy with a poor prognosis whose treatment has not changed significantly in 40 years.
In the past years, we have shown that SUMOylation controls AML cells response to chemotherapeutic drugs (anthracyclines such as daunorubicin or idarubicin and nucleoside analogues such as aracytin) used in standard AML treatments (Bossis et al., Cell Reports, 2014) as well as to differentiation therapies using All-trans-retinoic acid (Baik et al., Cancer Research 2018). In addition, we have shown that deregulation of SUMOylation and ubiquitylation enzyme activity is associated with AML resistance to standard chemotherapies (Gâtel et al., Life Science Alliance, 2020) and might serve as a biomarker of AML response to these treatments (patent EP19305688, 2019).
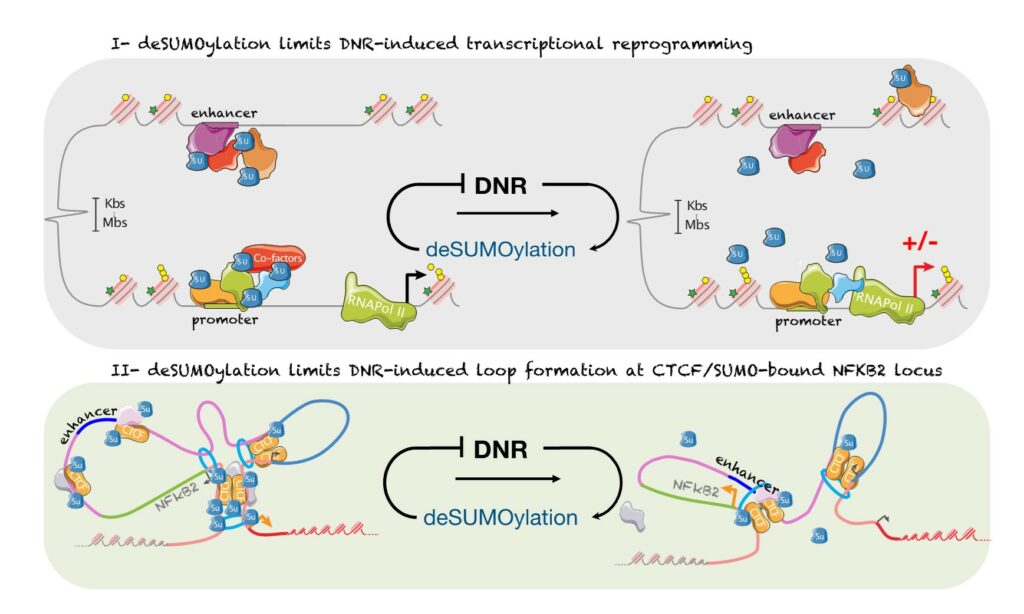
SUMOylation controls the rapid transcriptional reprogramming induces by chemotherapies in AML
We have shown that daunorubicin, the main chemotherapeutic drug used for AML induces a rapid transcriptional reprogramming in leukemic cells with almost 1000 genes being up- or down-regulated in less than 3 hours. This reprogramming is preceded by a massive deconjugation of SUMO from its target proteins, in particular those bound to promoters and enhancers. This deSUMOylation limits DNR-induced transcriptional reprogramming. We then studied in more details, NFKB2, one of the top DNR-induced genes. Our data suggest that DNR induces the formation of a CTCF-mediated loop between its promoter and a distal enhancer, which could explain NFKB2 induction. Finally, inhibition of SUMOylation prevent the formation of the loop and the induction of NFKB2 gene. Hence, our work shed new lights on the mode of action of chemotherapeutic drugs (Boulanger et al., Nucleic Acids Research, 2023)
SUMOylation inhibitor TAK-981 synergizes with Azacitidine in preclinical models of Acute Myeloid Leukemias and limits immune escape
The recent discovery of TAK-981 (Subasumstat), a first-in-class SUMOylation inhibitor, has allowed us to test in preclinical models the therapeutic relevance of targeting the SUMO pathway in AML. We could show that TAK-981 is endowed with potent anti-leukemic activity in various preclinical models of AML. TAK-981 targets AML cell lines and patient blast cells in vitro and in vivo in xenografted mice with minimal toxicity on normal cells. Moreover, it synergizes with 5-azacitidine (AZA), a DNA-hypomethylating agent now used in combination with the BCL-2 inhibitor venetoclax to treat patients unfit for standard chemotherapies. Interestingly, TAK-981+AZA combination shows higher anti-leukemic activity than AZA+venetoclax combination in vivo, at least in the model tested. Mechanistically, TAK-981 potentiates the transcriptional reprogramming induced by AZA, promoting differentiation and death of the leukemic cells. In addition, TAK-981+AZA treatment induces many genes linked to inflammation and immune response pathways. In particular, it leads to the secretion of type I interferon (IFN-I) by AML cells. Finally, TAK-981+AZA induces the expression of Natural Killer (NK)-activating ligands (MICA/B) and adhesion proteins (ICAM-1) at the surface of AML cells. Consistently, TAK-981+AZA-treated AML cells activate NKs and increase their cytotoxic activity. Targeting SUMOylation with TAK-981 may thus be a promising strategy to both sensitize AML cells to AZA and reduce their immune-escape capacities (Gabellier et al, Haematologica, 2023).
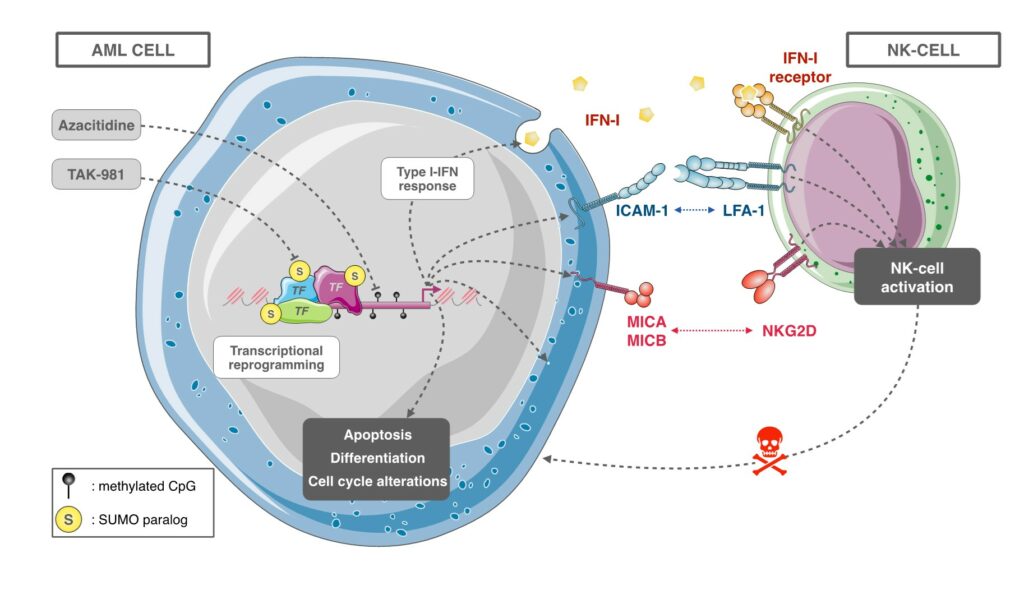
Our current work at better understanding, at the molecular level, the role of SUMOylation in (ii) the response of AMLs to therapies (chemotherapies, differentiation therapies and epidrug-based therapies), (ii) the activation of an anti-tumor immune response, in particular through the activation of Natural Killer cells. We also aim to further validate, in preclinical models of AMLs, the efficacy of SUMOylation targeting in the treatment of AMLs, alone or in combination with existing therapies. This work is carried out in close collaboration with the Department of Clinical Hematology of the University Hospital of Montpellier. Finally, we are developing new tools and molecules to target SUMOylation in AML and more generally cancer, in particular through our ongoing collaboration with Dr Muriel Amblard’s team (IBMM).