The R2TP chaperone recruits HSP90 to assemble numerous cellular nanomachines, including RNA polymerases and PIKKs regulatory kinases. In the gut, this process occurs in proliferating cells and is essential for tissue homeostasis. When inactivated, this leads to cycle arrest and cell death. Furthermore, R2TP is a poor prognostic factor in patients with colorectal cancer. This work, published in the journal Nature Communications, shows for the first time links between protein complex assembly and normal or pathological cell growth.
The expression of genes into functional entities relies on the transcription of genes into mRNA, their translation into proteins, their folding into their final form, and, very often, their incorporation into functional macromolecular complexes. Indeed, most proteins are only active once integrated into complexes containing several subunits, which then function as nanomachines. This is notably the case of RNA polymerases, responsible for the transcription of DNA into RNA, ribosomes, which decode mRNA into polypeptide chains, and TORC1 and TORC2 complexes including mTOR, which regulate protein synthesis according to the nutritional state of the cell. While the activity of these nanomachines has been extensively studied, their assembly from individual subunits is still poorly understood.
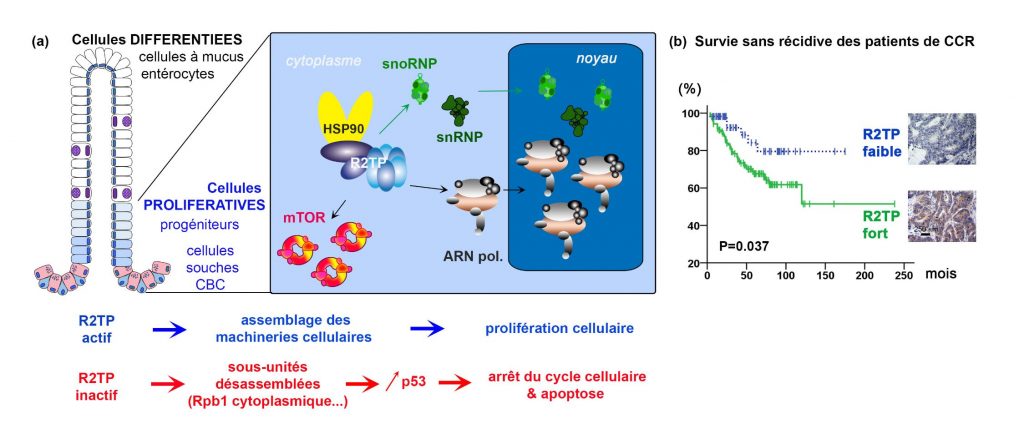
HSP90 is a chaperone involved in the folding of many substrates. The activity of HSP90 involves different adapters, each specialized for a batch of substrates. Over the past 15 years, a new adapter, named R2TP, has been identified. R2TP was first discovered for its role in the construction of snoRNPs, small particles of protein and non-coding RNA that ensure ribosome maturation. Subsequently, R2TP was found to assemble other essential nanomachines in the cell, including (a) other non-coding RNA-containing particles (snRNPs, miRNPs), (b) RNA polymerases responsible for the transcription of DNA into RNA, (c) functional complexes of mTOR, whose kinase activity regulates cell growth and proliferation, and (d) complexes of ATR, ATM and DNA-PKcs, kinases that signal and repair DNA damage. In fact, the list of R2TP substrates is growing, so that additional substrates are likely to be discovered. R2TP would mobilize HSP90 activity to complete the folding of substrate proteins and allow their incorporation into active nanomachines. However, the underlying assembly mechanisms remain poorly understood, as does the role of R2TP in tissues and organisms.
Under the lead of Berengere Pradet Balade (CRBM) and involving the Hahne team of the IGMM four teams in Montpellier collaborated to address this issue. Together, we could show that R2TP is essential for tissue homeostasis. Murine models of conditional invalidation in the intestinal epithelium have shown the essential role of R2TP in the maintenance of epithelial homeostasis. Indeed, R2TP is essential for the proliferation of intestinal stem and progenitor cells. Its inactivation induces signaling pathways that converge towards cycle arrest and cell death of this proliferative compartment. As a result, the intestinal epithelium, one of the most rapidly renewing tissues in the body, cannot renew itself and degenerates. In fact, the sensitivity of tissues to R2TP inactivation correlates with the speed of tissue turnover: the murine small intestine, renewed in 3-4 days, degenerates before the colon, which renews more slowly. Moreover, R2TP inactivation prevents the formation of intestinal and colonic organoids in vitro, with kinetics that reflect histological observations. These data demonstrate the essential role of the R2TP chaperone and its assembly function in intestinal cell proliferation.
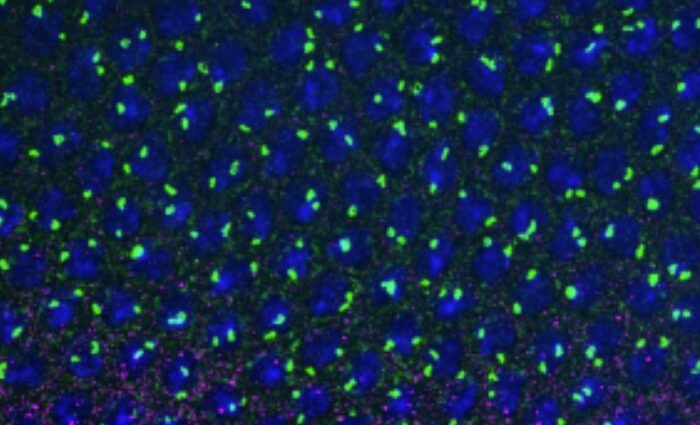
In histological sections of intestine, inactivation of R2TP causes cytoplasmic accumulation of Rpb1, the catalytic subunit of RNA polymerase II. This accumulation is visible in stem cells and intestinal progenitors, but not in differentiated cells. This accumulation reflects the inability of Rpb1 to assemble with its partners into a functional RNA polymerase, which would then be addressed to the nucleus. This phenotype coincides with activation of the p53 pathway and cell cycle arrest. The p53 protein is known, among other things, to be a sensor of defects in ribosome production, via direct interaction with unincorporated ribosomal proteins. An intriguing possibility, yet to be demonstrated, is that p53 also functions as a sensor of the assembly of other cellular nanomachines.
Finally, a correlation was observed between disease free survival and R2TP levels in patients with colorectal cancer. R2TP thus appears to be a poor prognostic factor in these patients.
For more information:
The HSP90/R2TP assembly chaperone promotes cell proliferation in the intestinal epithelium
Maurizy C, Abeza C, Lemmers B, Gabola M, Longobardi C, Pinet V, Ferrand M, Paul C, Bremond J, Langa F, Gerbe F, Jay P, Verheggen C, Tinari N, Helmlinger D, Lattanzio R, Bertrand E, Hahne M, Pradet-Balade B.
Nature Communications. 10 août 2021. doi: 10.1038/s41467-021-24792-4.
Scientists contact :
Bérengère PRADET-BALADE Chercheuse CNRS pradet@crbm.cnrs.fr (+33) 4 34 35 95 48 CRBM (CNRS/UM) 1919, route de Mende 34293 Montpellier Cedex 05
Michael HAHNE Directeur de recherche Inserm michael.hahne@igmm.cnrs.fr (+33) 4 34 35 96 39 IGMM (CNRS/UM) 1919, route de Mende 34293 Montpellier Cedex 05
Edouard BERTRAND Directeur de recherche CNRS edouard.bertrand@igh.cnrs.fr (+33) 4 34 35 99 21 IGH (CNRS/UM) 141, rue de la Cardonille 34094 Montpellier Cedex 5