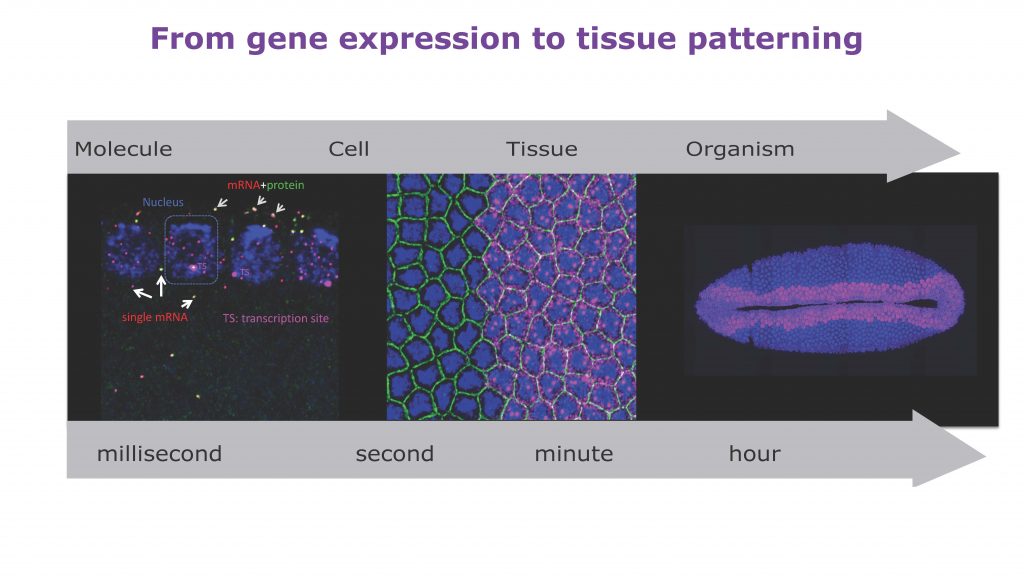
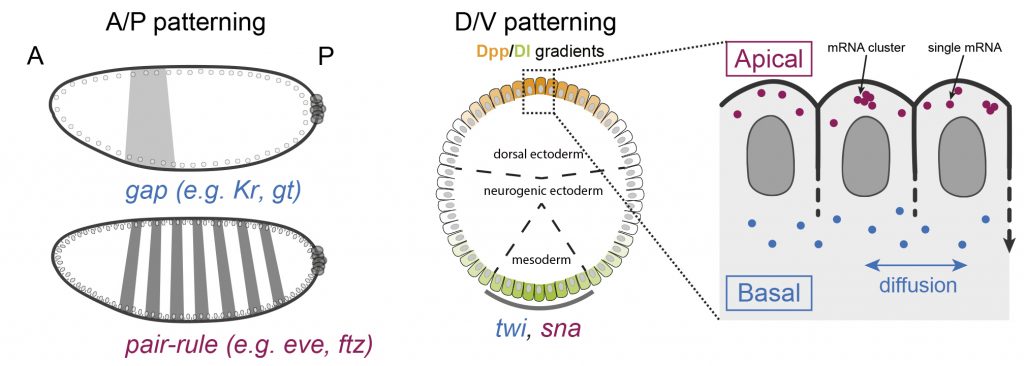
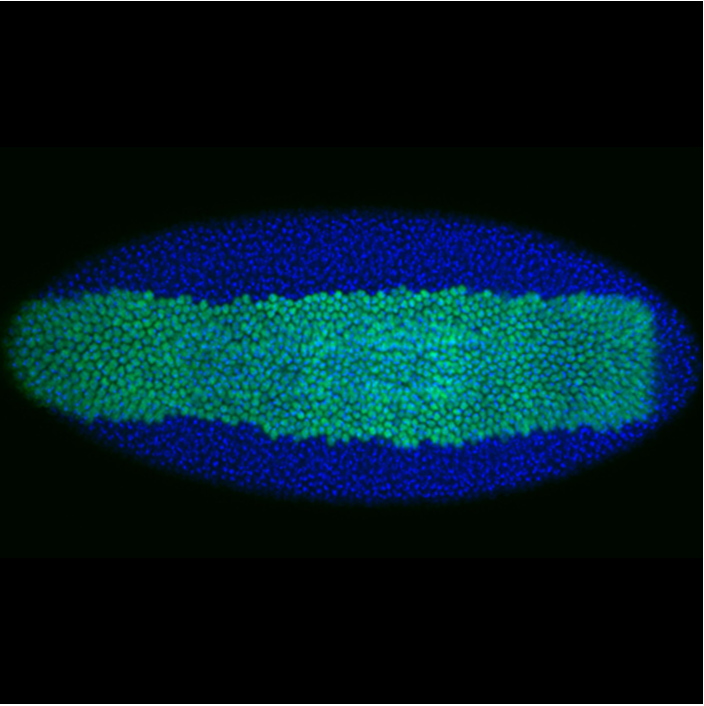
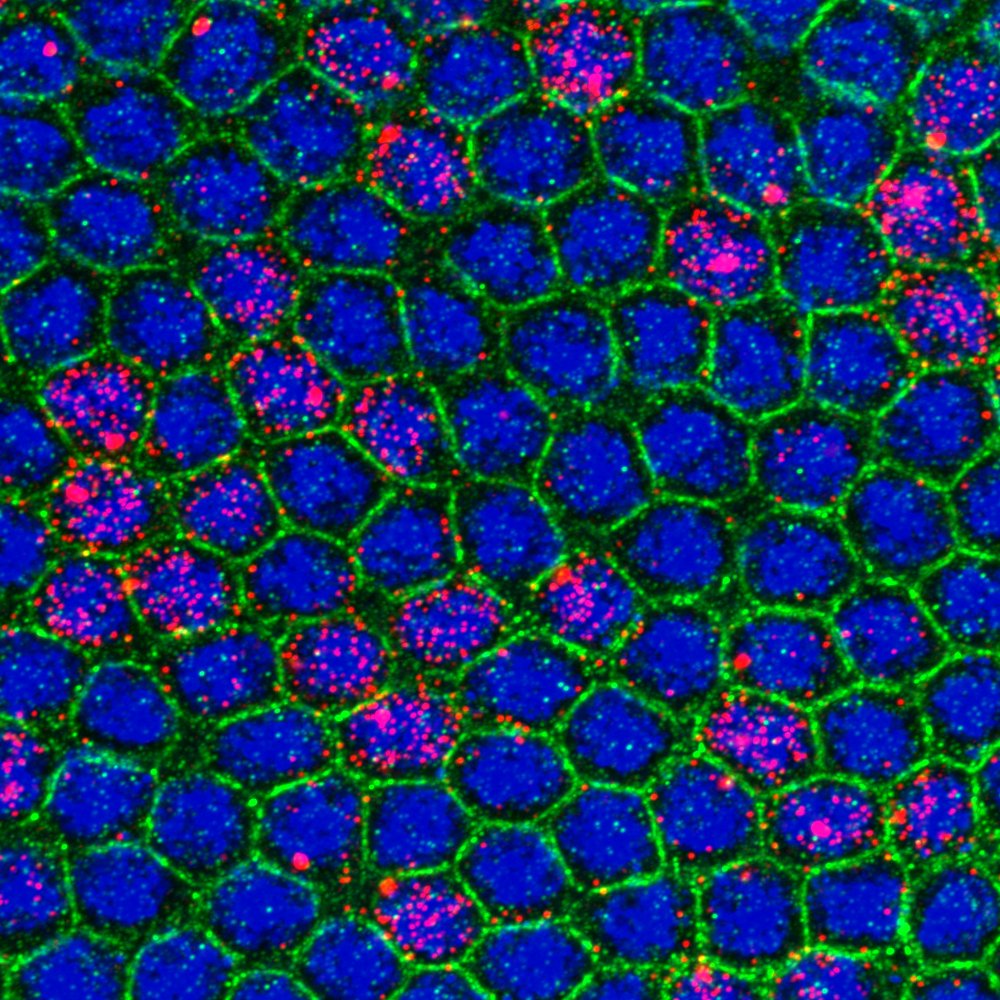
During development, precise control of gene expression allows the reproducible establishment of patterns, leading to the formation of organs at the right time and place. What are the mechanisms behind such precision?
The establishment of developmental patterns has been primarily studied at the transcriptional level. In comparison, the fate of these transcripts received little attention. Development of new methods enabling the study of translation of single mRNA molecules revealed a critical heterogeneity in translation. Our lab pioneered the deployment of such methods in Drosophila embryos, leading to the discovery of translation factories and unmasking intragenic translation heterogeneity, demonstrating a novel layer of variation in gene expression. Thus, to decipher the mechanisms dictating precision in gene expression, it is essential to simultaneously consider both layers of regulation constituted by transcription and translation as well as their potential coupling.
We propose to bridge the gap between these two pillars of the central dogma to obtain an integrated multiscale view of gene expression control. We use quantitative imaging methods to simultaneously monitor mRNA and protein in living Drosophila embryos with unprecedented spatio-temporal resolution, combined with theoretical models to formulate predictions that will be tested by genetic/optogenetic manipulations.
PROJECT 1
How do translation dynamics of localized patterning transcripts modulate cell fate decisions?