Our group studies epigenetics in mammals and we are particularly interested in the epigenetic mechanisms that control imprinted genes. These rather unusual genes are organized in evolutionarily conserved chromosomal domains and play diverse but essential roles in development and disease. Imprinted genes are expressed in a mono-allelic manner, depending entirely on whether the gene is inherited from the mother or from the father. This unusual allelic gene expression is controlled by differential DNA methylation ‘imprints’ at key regulatory sequences, and other features of chromatin are critically involved as well.
One theme of the lab is to explore the germline establishment and the somatic maintenance of these epigenetic imprints. Tightly linked to this research question, is the issue of how the methylation imprints bring about imprinted gene expression in the embryo, often in a tissue-specific manner. We have explored this question for selected imprinted domains and our ongoing studies have pinpointed the critical involvement of histone lysine methylation and long non-coding RNAs.
During the last years, our lab has investigated a second theme related to epigenetics, namely the structuration of chromosomal domains within the nucleus and how this can be explored using new technologies. One question which we have investigated for selected imprinted domains, is whether chromatin is structured differently on the maternal versus paternal chromosomes, and whether such differential structuration contributes to the allelic expression of imprinted genes. In parallel, we have used advanced fluorescence microscopy approach to monitor nanoscale chromatin compaction and structuration in living cells. Together with gene targeting approaches, this novel FRET-based assay performed in mouse and nematode model systems allow one to unravel the structural roles of candidate chromatin regulators. The combined studies of our lab aim to enhance our mechanistic understanding of chromatin and imprinted gene regulation, and how this can contribute studying specific human diseases
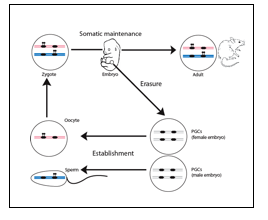
Epigenetic mechanisms play diverse roles in development and mediate stable repression or expression of genes in specific cell lineages. These diverse mechanisms can be perturbed by environmental and toxic components, sometimes with long-lasting consequences. In mammals, there are several examples of somatically heritable inactivation of one of the two alleles of genes. These include genes on the X-chromosome in females (‘X-inactivation’) and imprinted genes, an essential set of ~150 protein-coding genes (and hundreds of regulatory non-coding RNA genes). Imprinted gene expression depends entirely on whether the gene is inherited from the mother or the father, and these exceptional genes play key roles in cell proliferation, homeostasis and behaviour. The parent-of-origin specific, mono-allelic, expression of imprinted genes is mediated by so-called imprinting control regions (ICRs). These are essential GC-rich sequence elements of several kilobases onto which epigenetic marks are placed depending on their passage through either the female or the male germ line. The precise nature of these parental epigenetic imprints is not yet fully understood, but they are characterized by DNA methylation. The male- and female-specific establishment of DNA methylation imprints in the germ cells requires the de novo methyltransferase Dnmt3a and its co-factor Dnmt3-like (Dnmt3l). After fertilization, exceptionally, the maternal and paternal methylation imprints are maintained throughout development, in all the somatic cells (Figure 1). Deregulation of this essential process is causally involved in the foetal growth-related syndromes Silver-Russell Syndrome (SRS), Beckwith-Wiedemann Syndrome (BWS) and Transient Neonatal Diabetes Mellitus (TNDM), as well as in different neuro-behavioural syndromes.
Similarly as at tumour suppressor genes, DNA methylation at imprinted loci is often perturbed in cancer and this contributes to tumorigenesis.
Figure 1: Developmental control of the DNA methylation that controls the allelic expression of imprinted genes. Hypothetical methylation imprints (black lollipops) are shown. The maternal genome is depicted in pink, the paternal one in blue. In Primordial germ cell (PGCs), the imprints are erased to allow a new ‘cycle of imprinting’ for the next generation.
Adapted from Hirasawa & Feil, BioEssays 2010.
1.Maintenance of parental methylation imprints during development.
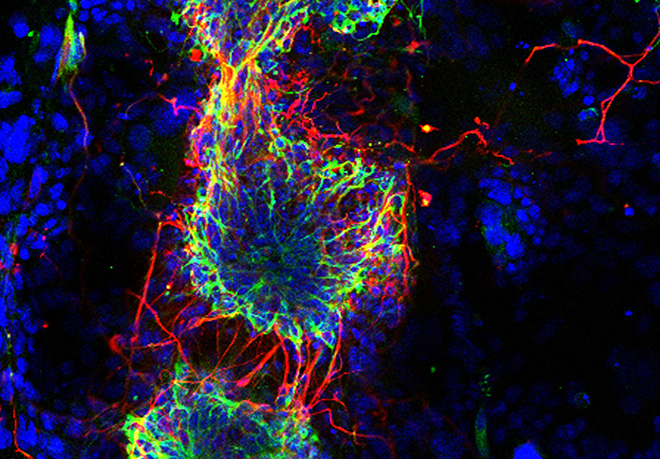
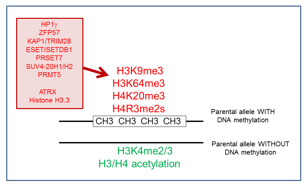
Within this theme, we focus on the mono-allelic DNA methylation imprints at ‘imprinting control regions’ (ICRs) and how these are maintained during development, particularly during the critical pre-implantation stages. The team has explored the presence and importance of histone lysine and arginine methylation at the chromatin associated with ICRs, which has a very different combination of marks on the DNA-methylated allele compared to the unmethylated allele (Figure 2). More recently, we have become interested in the role of specific transcription factors. Studies use the mouse as model system and explore different imprinted domains and genome-wide aspects of epigenetic regulation. Linked to the exploration of chromatin at ICRs and how their differential DNA methylation is regulated throughout development, is the interesting question about their evolutionary origin and relevance. This theme is of particularly relevant also for human diseases in which losses, or gains, of DNA methylation are causally involved. Therefore, we have established collaborations with clinical groups to explore the conservation of the underlying mechanisms in humans and to explore their importance in imprinting disorders.
Recent publications on this research theme
Henckel A., Chebli K., Kota S.K., Arnaud P., and Feil R. (2012). Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO Journal, 31, 606-615.
Girardot M., Hirasawa, R., Kacem, S., Fritsch, L., Pontis, J., Kota, S.K., Filipponi, D., Fabbrizio, E., Sardet, C., Lohmann, F., Kadam, S., Ait-Si-Ali, S. and Feil, R. (2014). PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G+C rich regions of the mouse genome. Nucleic Acids Research, 42, 235-248.
Auclair, G., Borgel, J., Sanz, L.A., Vallet, J., Guibert, S., Dumas, M., Cavelier, P., Girardot, M., Forné, T., Feil, R.* and Weber, M.* (2016). EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res, 26, 192-202.
Prats-Puig, A., Carreras-Badoso, G., Bassols, J., Cavelier, P., Magret, A., Sabench, C., de Zegher, F., Ibanez, L., Feil, R.*, Lopez-Bermejo, A.* (2017). The placental imprinted DLK1-DIO3 domain: a new link to pre- and postnatal growth in human beings. Am. J. Obstet. & Gynecology, Epub before print.
Figure 2: Differential chromatin organization at ‘imprinting control regions’ (ICRS) in somatic cells. On the parental allele marked by the DNA methylation imprint, there is repressive histone methylation (in red) regulated by different proteins (shown to the left). The opposite, non-methylated allele is marked by ‘active chromatin’ modifications (in green).
(Sanli et al., Int J. Biochem. Cell Biol. 2015).
2.How do ICRs bring about imprinted gene expression in cis?
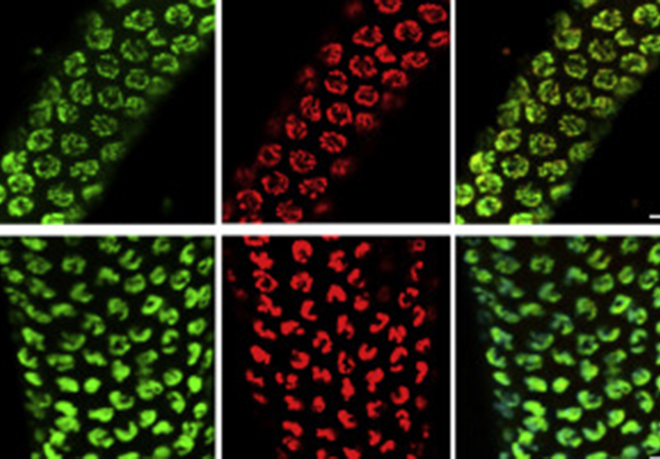
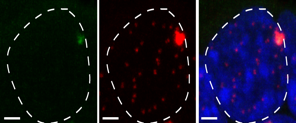
This has been a long-standing theme of the laboratory and our research addresses how the parentally-acquired DNA methylation marks at CpG islands bring about the mono-allelic gene expression at imprinted chromosomal domains. This poorly understood process occurs during embryogenesis, often in a lineage-specific manner. In earlier work, we showed the importance of lysine methylation at histones in the developmental control of imprinted gene expression. Particularly, we reported diverse roles of the H3 lysine-9 methyltransferase G9A (EHMT2) and that of the type-2 Polycomb Repressive Complex (PRC-2). At several imprinted domains, the ICR produces long ncRNAs that are retained in the nucleus and alter gene expression in cis. To further address their mechanisms of action, we have studied the Dlk1-Dio3 imprinted domain on mouse chromosome 12. We found that the intergenic ICR of this 1-Mb domain acts as an enhancer, controlling in cis the allelic expression of multiple ncRNAs. One of these maternally activated ncRNAs is suspected to repress in cis the protein-coding genes of this large domain domain, a process which we have been exploring in more detail. Interestingly, this long ncRNA, called Meg3 (Maternally expressed gene-3), has the unusual feature that it is retained at its site of transcription. We have developed different ways to visualize the Meg3 lncRNA molecules in the nucleus (Figure 3) based on MS2-tagging approach and single molecule RNA FISH detection. We are performing functional studies to explore its role in gene regulation in embryonic stem cells and neural progenitor cell lines.
Recent publications on this research theme
Kota, S.K.*, Lleres, D.*, Bouschet, T., Hirasawa, R., Marchand, A., Begon-Pescia, C., Sanli, I, Arnaud, P., Journot, L., Girardot, M. and Feil, R. (2014). ICR non-coding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev Cell, 31, 19-33.
Puget, N., Hirasawa, R., Nguyen Hu, N-S., Laviolette-Malirat, N., Feil, R.* & Khamlichi, A.A.* (2015). Insertion of an imprinted insulator into the IgH locus reveals developmentally regulated, transcription-dependent suppression of V(D)J recombination. Mol Cell Biol. 35, 529-543.
Figure 3: FISH detection of the Meg3 long non-coding RNA of the Dlk1-Dio3 imprinted domain in ES cells, using a classical cDNA probe (green) or fluorescent oligonucleotides that detect the inserted MS2 repeats (red). The image to the right shows the overlay. Scale bar, 5 mm.
(Sanli/Lalevée et al., Cell reports 2018)
3.Structural organisation of imprinted gene domains in the nucleus.
It remains poorly understood how imprinted chromosomal domains are organised in the 3D space of the nucleus, and whether their structural organisation is different on the maternal compared to the paternal genome. A related question is whether the higher-order organisation(s) of imprinted domains is developmentally regulated, and could thus contribute to tissue-specific imprinted gene expression. To explore the nuclear topology of chromatin domains, we have chosen the Dlk1-Dio3 locus on mouse chromosome-12 as a model locus and also other imprinted domains. In our studies on embryonic stem (ES) and differentiated cells we use different DNA Fluorescence in-situ hybridization (FISH) approaches and, in collaboration with others, apply chromosome conformation capture (4C) techniques. Combined, these approaches provide important insights into the differential structuration on the maternal versus paternal chromosomes, and whether such differential chromatin organization contributes to the allelic expression of imprinted genes.
Recent publications on this research theme
Kota, S.K.*, Lleres, D.*, Bouschet, T., Hirasawa, R., Marchand, A., Begon-Pescia, C., Sanli, I, Arnaud, P., Journot, L., Girardot, M. and Feil, R. (2014). ICR non-coding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev Cell, 31, 19-33.
Sanli, I. and Feil, R. (2016). PRC-mediated interaction networks of repressed genes: emerging insights and possible roles. Epigenomics, 8, 733-735.
(see also Kota/Llères et al. Developmental Cell 2014).
4.Exploration of nanoscale chromatin structuration in living cells and animals
Recently, we have adapted a FRET-based microscopy approach to assess compaction of chromatin in living cells. In particular, this novel approach, based on the non-invasive fluorescence lifetime imaging (FLIM) technique, monitors the nanoscale structure of chromatin and its compaction state in living conditions. We successfully used this quantitative microscopy approach in living nematodes, to define chromatin compaction states along meiotic chromosomes, and the role of condensin complexes at specific meiotic stages. This nanoscale chromatin compaction assay works well in human cancer cells as well, which has allowed us to explore the chromatin structural roles of specific chromatin associated-proteins. For the future, we aim to get this experimental system to work in primary mouse cells, to then explore the role of chromatin modifiers in the nanoscale chromatin structuration of chromatin and it offers the exciting prospect to explore the effects of genetic and environmental factors on chromatin compaction.
Recent publications on this research theme
Lleres, D.*, Bailly, A.P.*, Perrin, A., Norman, D., Xirodimas, D.P., Feil, R. (2017). Quantitative FLIM-FRET microscopy to monitor nanoscale chromatin compaction in vivo reveals structural roles of condensin complexes. Cell reports, 18, 1791-1803.
Sobecki M, Camasses A, Mrouj K, Parisis N, Nicolas E, Lleres D, Gerbe F, Prieto S, Krasinska L, David A, Egure M, Birling M-C, Hem S, Dejardin J, Malumbres M, Jay P, Dulic V, Lafontaine DLJ, Feil R, Fisher D (2016). Ki-67 organises heterochromatin and controls gene expression during cell proliferation. ELife, e13722.